Copyright
©The Author(s) 2022.
World J Gastroenterol. Aug 7, 2022; 28(29): 3886-3902
Published online Aug 7, 2022. doi: 10.3748/wjg.v28.i29.3886
Published online Aug 7, 2022. doi: 10.3748/wjg.v28.i29.3886
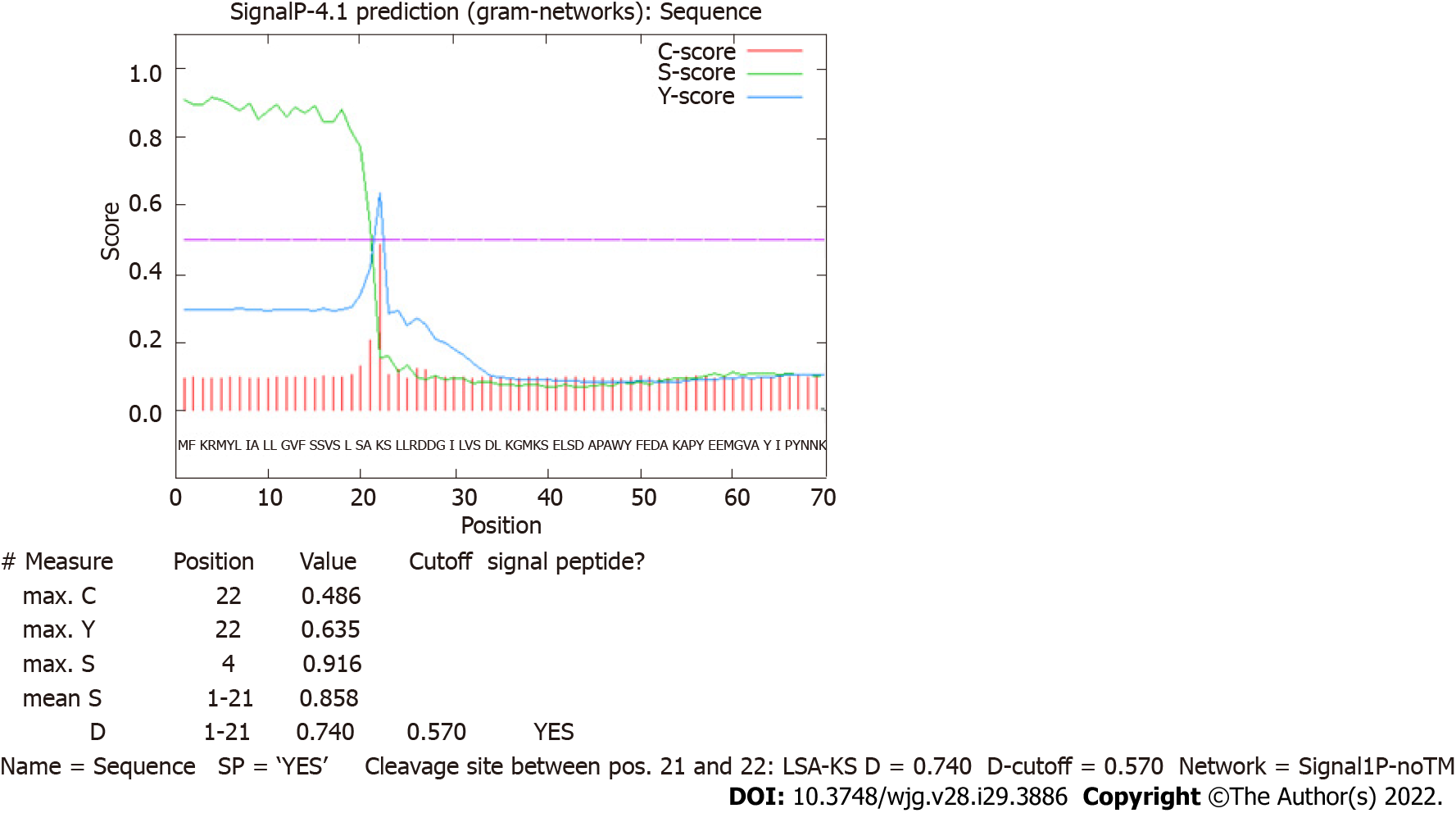
Figure 1 Prediction of a signal peptide for HP0953 protein.
The green line that represents the S-score indicates a high probability that the first 20 amino acids conform a signal peptide that can be removed by a cut between amino acids 20 and 21, as marked by the tallest red line indicating the C-score.
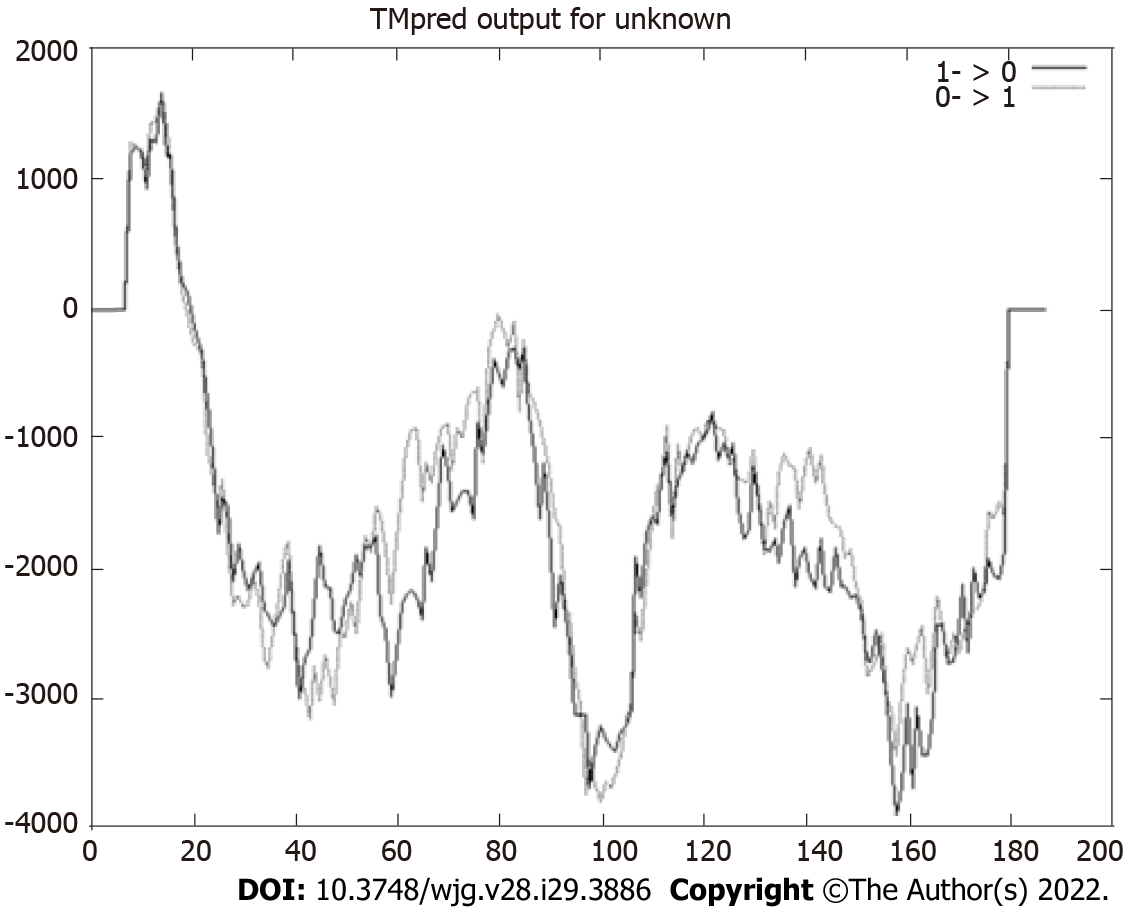
Figure 2 Prediction of transmembrane helices.
Values > 0 indicate the probable presence of transmembrane helices, as observed in the region constituted by the first 25 amino acids. The HP0953 protein could be anchored to the membrane to be subsequently secreted.
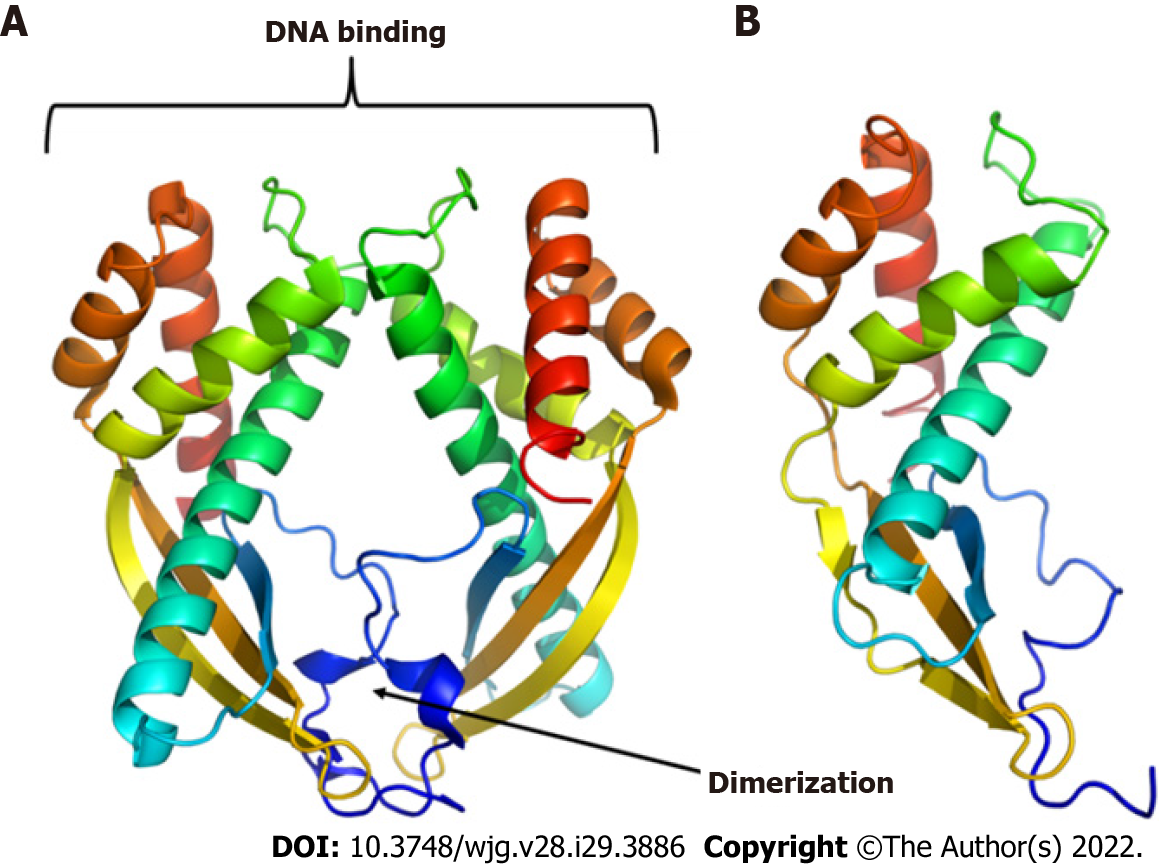
Figure 3 Three-dimensional models of proteins.
A: Tip-alpha protein from Helicobacter pylori (H. pylori); B: HP0953 protein from H. pylori. Tip-alpha acquires a dimer arrangement that could also be present in the HP0953 protein due to its structural similarity in the position of the alpha helices and beta sheets.
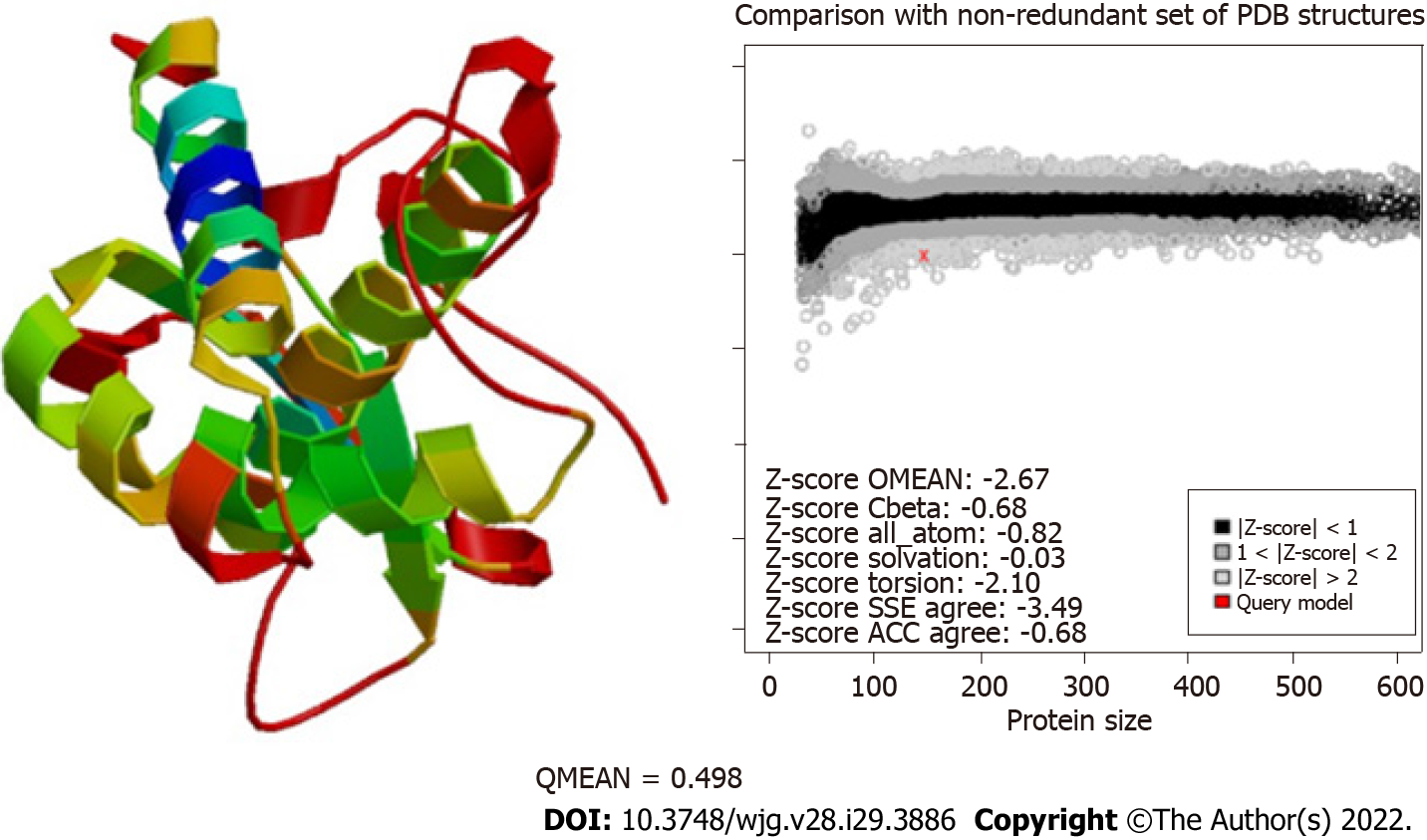
Figure 4 Evaluation of the modeling quality of HP0953.
The QMEAN for this protein was 0.498, which indicates that the obtained model is acceptable. PDB: Protein Data Bank; SSE: Secondary Structure Element; ACC: Solvent Accessibility.
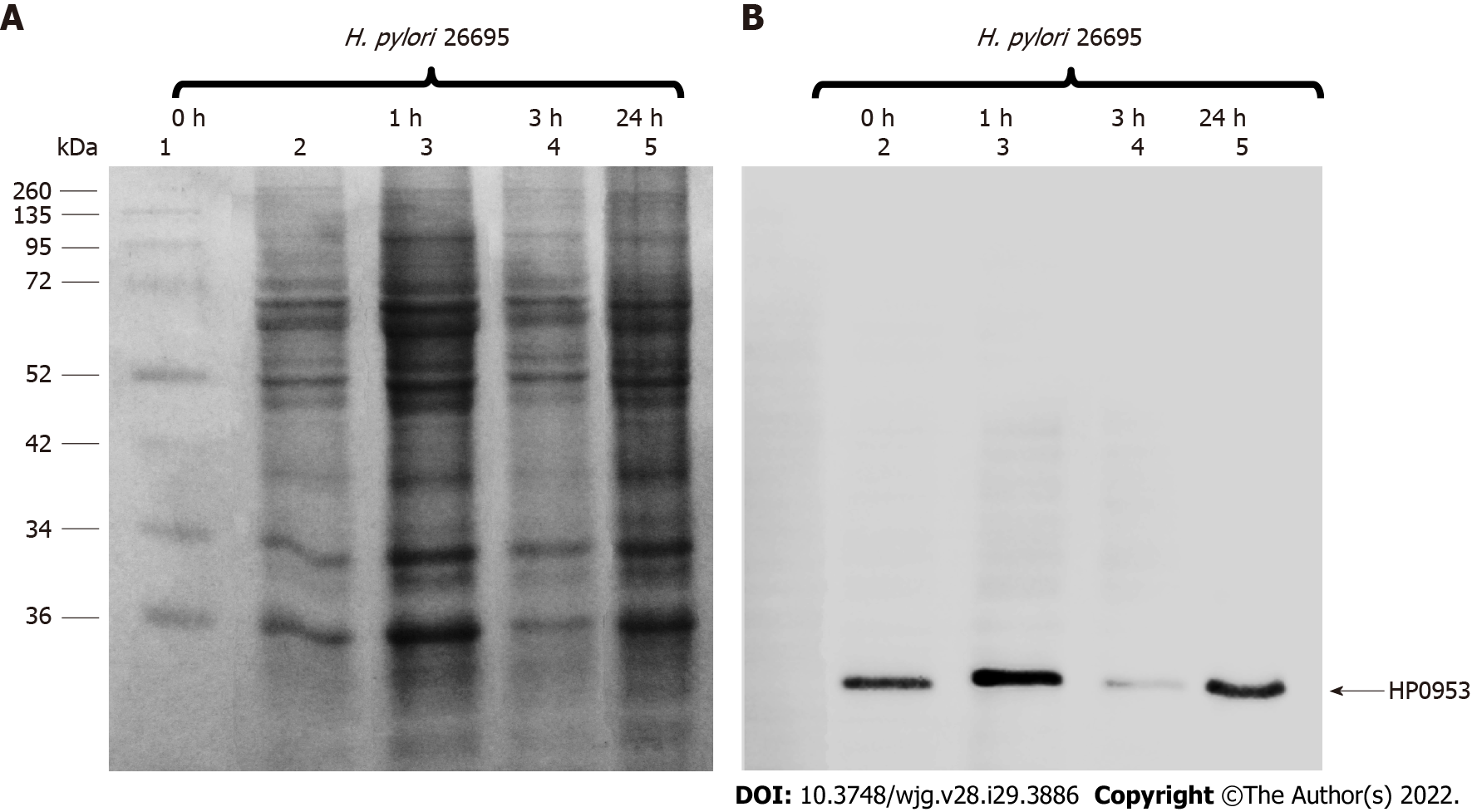
Figure 5 Location of the hypothetical protein HP0953 in the insoluble fraction of Helicobacter pylori 26695.
A: Sulfate-polyacrylamide gel electrophoresis 16% gel stained with Coomassie blue; B: Western blot with an arrow showing the band corresponding to the expected molecular weight. Lanes: (1) Molecular weight marker and (2-5) insoluble fractions of Helicobacter pylori 26695 from samples collected at 0, 1, 3, and 24 h of incubation during biofilm formation. The arrow indicates the presence of the native HP0953 protein. H. pylori: Helicobacter pylori.
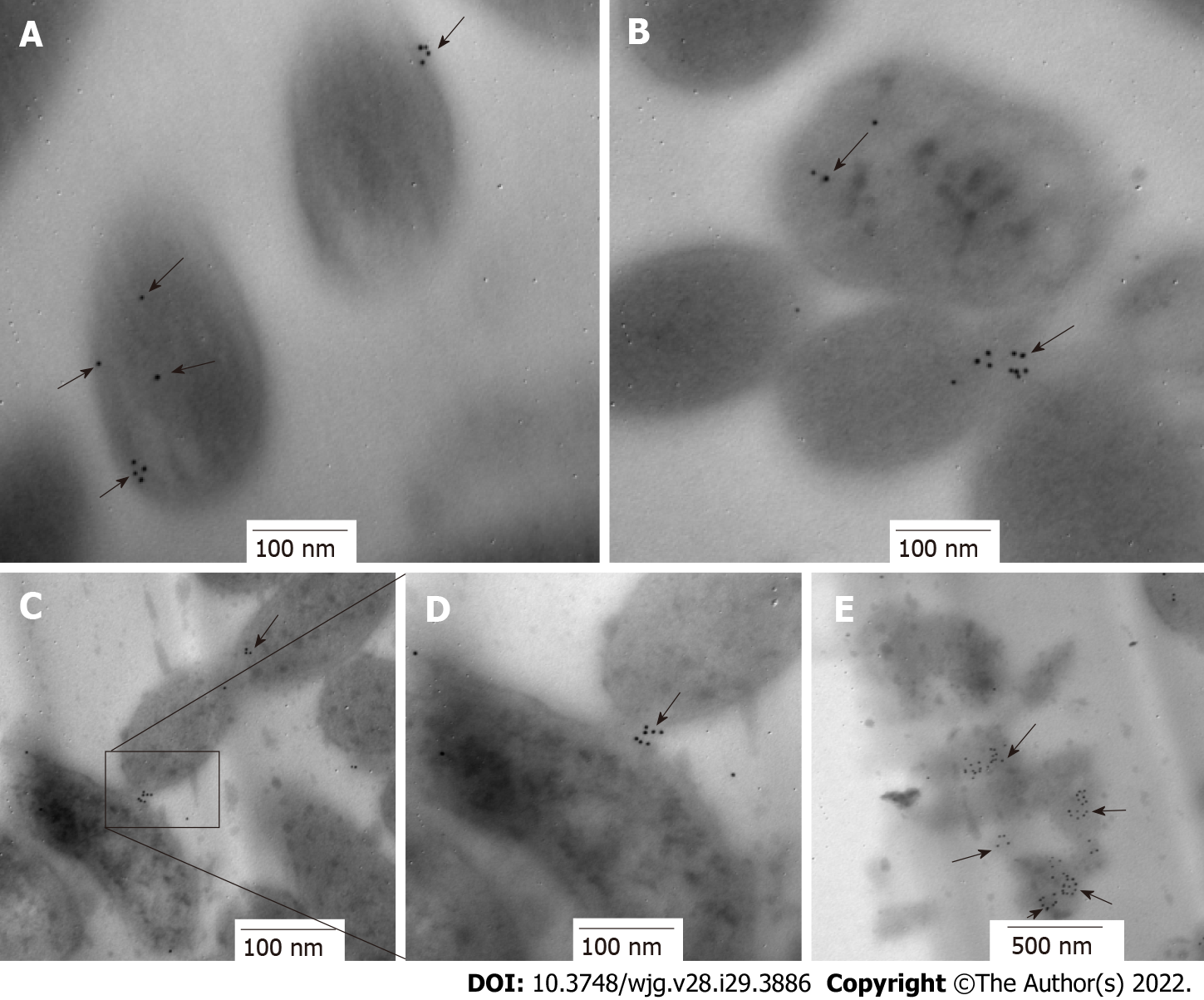
Figure 6 Immunolocation of HP0953 protein in Helicobacter pylori bacterial transmission electron microscopy sections.
A and B: 100000 ×; C: 75000 ×; D: 150000 ×; E: 60000 ×. The black arrows show the location of HP0953 protein. Immunogold technique, primary antibody (polyclonal rabbit anti-HP0953), secondary antibody (goat anti-rabbit immunoglobulin G coupled to 10-nm colloidal gold particles). (JEM 1010 JEOL Ltd. Tokyo, Japan) microscope equipped with a TEM Imaging Systems AMT (Woburn, MA, United States).
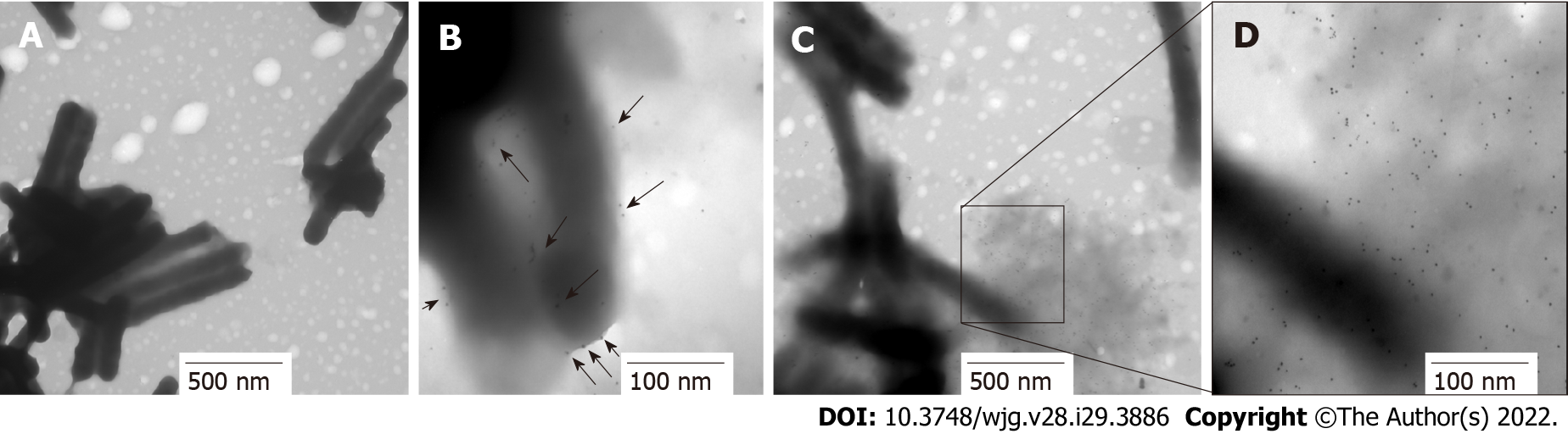
Figure 7 Immunolocation of HP0953 protein in whole bacteria.
A: Helicobacter pylori (H. pylori) incubated with the secondary antibody alone, 20000 ×; B-D: Complete immunogold technique for H. pylori; B: 100000 ×; C: 30000 ×; D: 100000 ×. Black arrows indicate the location of HP0953 protein. Immunogold technique, primary antibody (polyclonal rabbit anti-HP0953), secondary antibody (goat anti-rabbit immunoglobulin G coupled to 10-nm colloidal gold particles).
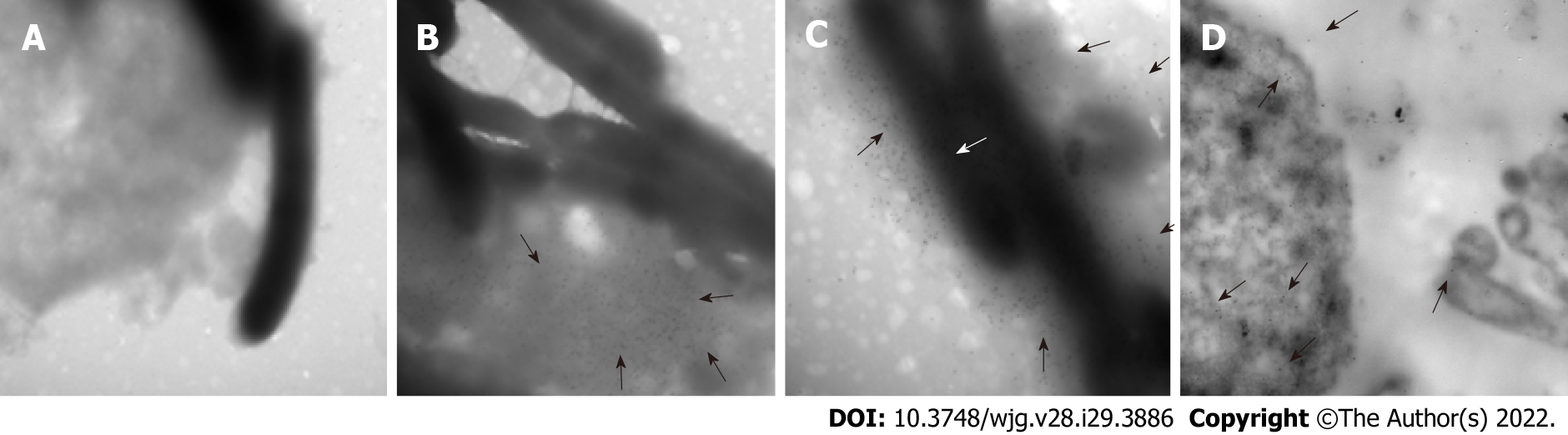
Figure 8 Location of HP0953 protein during against gastric cell infection by Helicobacter pylori.
A: Against gastric (AGS) cell at 3 h of infection by Helicobacter pylori (H. pylori), incubated with the secondary antibody alone, 40000 ×; B: Complete immunogold technique for AGS cells at 3 h of infection by H. pylori, 40000 ×; C: Complete immunogold technique for AGS cells at 3 h of infection by H. pylori, 50000 ×; D: Complete immunogold technique for AGS cells at 12 h of infection by H. pylori, 40000 ×. White arrows indicate the location of HP0953 protein.
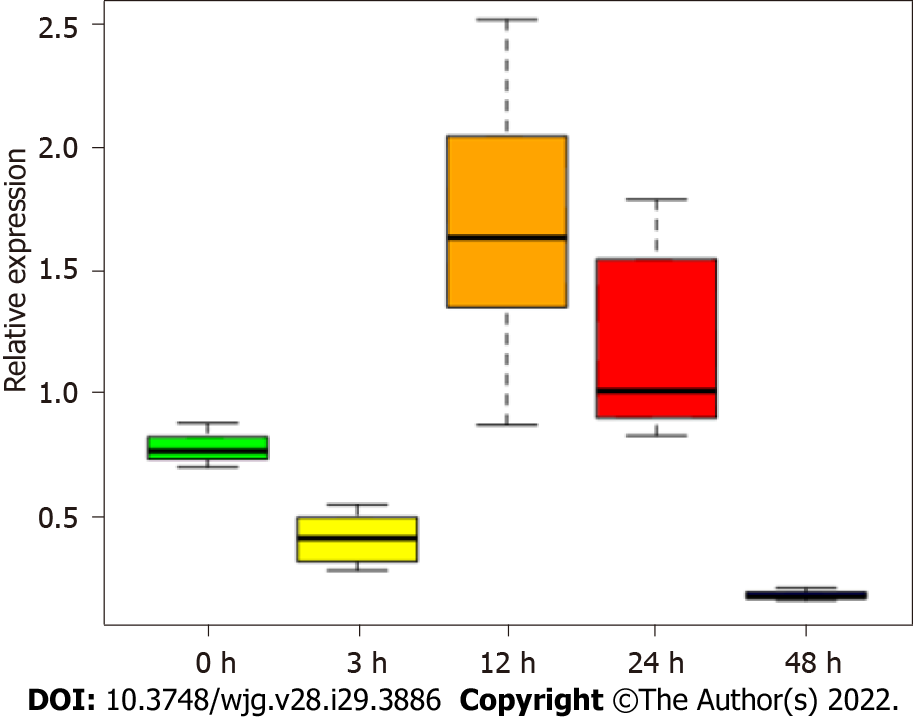
Figure 9 Expression of HP0953 increases significantly after 12 h of infection of against gastric cells.
The Kruskal-Wallis equality-of-populations rank test was used to compare the expression of the gene at all study time points. P = 0.0006.
- Citation: Arteaga-Resendiz NK, Rodea GE, Ribas-Aparicio RM, Olivares-Cervantes AL, Castelán-Vega JA, Olivares-Trejo JJ, Mendoza-Elizalde S, López-Villegas EO, Colín C, Aguilar-Rodea P, Reyes-López A, Salazar García M, Velázquez-Guadarrama N. HP0953 - hypothetical virulence factor overexpresion and localization during Helicobacter pylori infection of gastric epithelium. World J Gastroenterol 2022; 28(29): 3886-3902
- URL: https://www.wjgnet.com/1007-9327/full/v28/i29/3886.htm
- DOI: https://dx.doi.org/10.3748/wjg.v28.i29.3886