Copyright
©The Author(s) 2020.
World J Gastroenterol. Aug 7, 2020; 26(29): 4356-4371
Published online Aug 7, 2020. doi: 10.3748/wjg.v26.i29.4356
Published online Aug 7, 2020. doi: 10.3748/wjg.v26.i29.4356
Figure 1 Overview of a meta-analysis approach in this study.
CCA: Cholangiocarcinoma; DE: Differentially expressed; miRNA: MicroRNA.
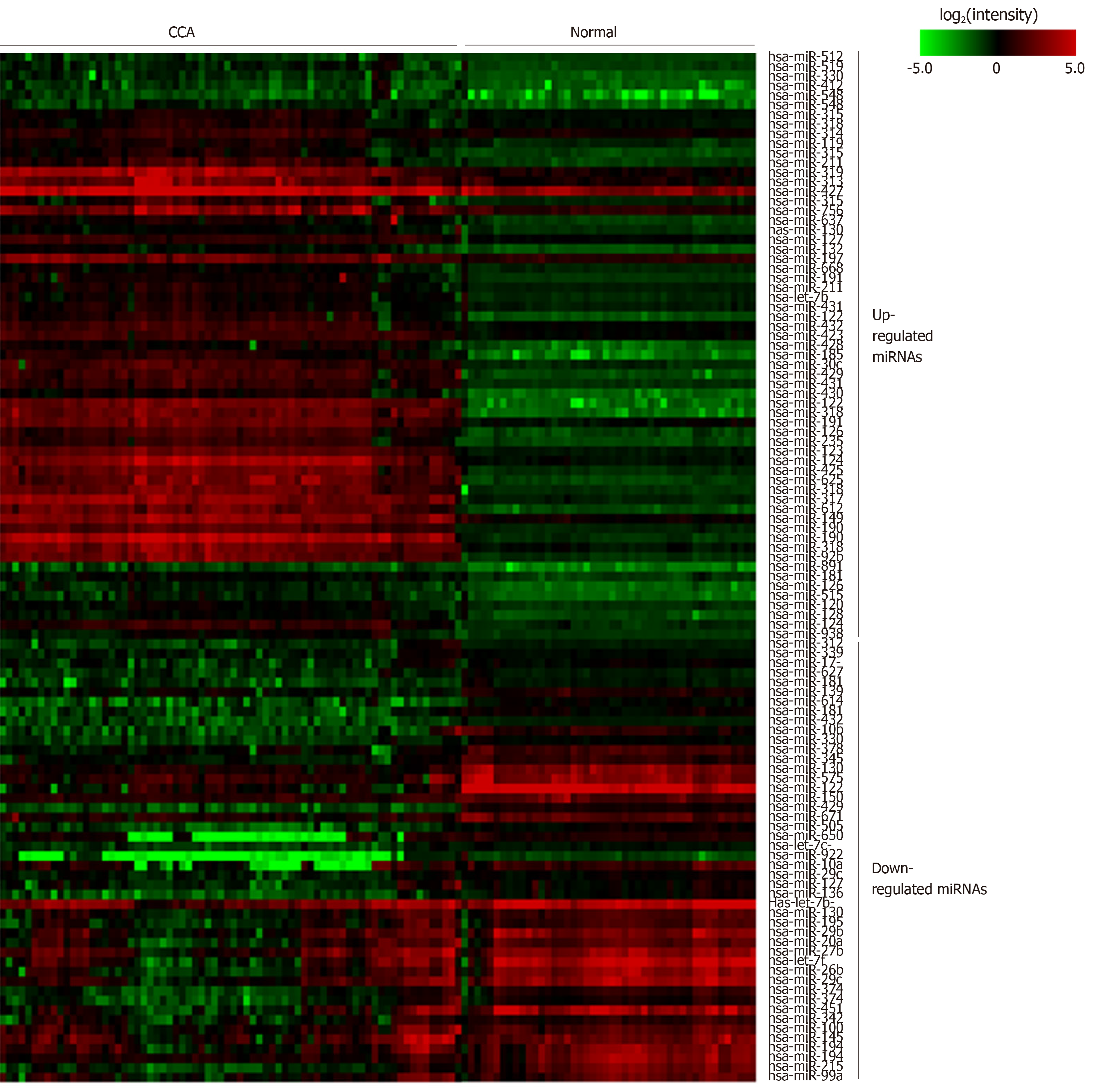
Figure 2 Heatmap of microRNA expression in cholangiocarcinoma and normal samples.
The color gradient of each cell represents the log2 of normalized intensity value of microRNA microarray spot. CCA: Cholangiocarcinoma; miRNA: MicroRNA.
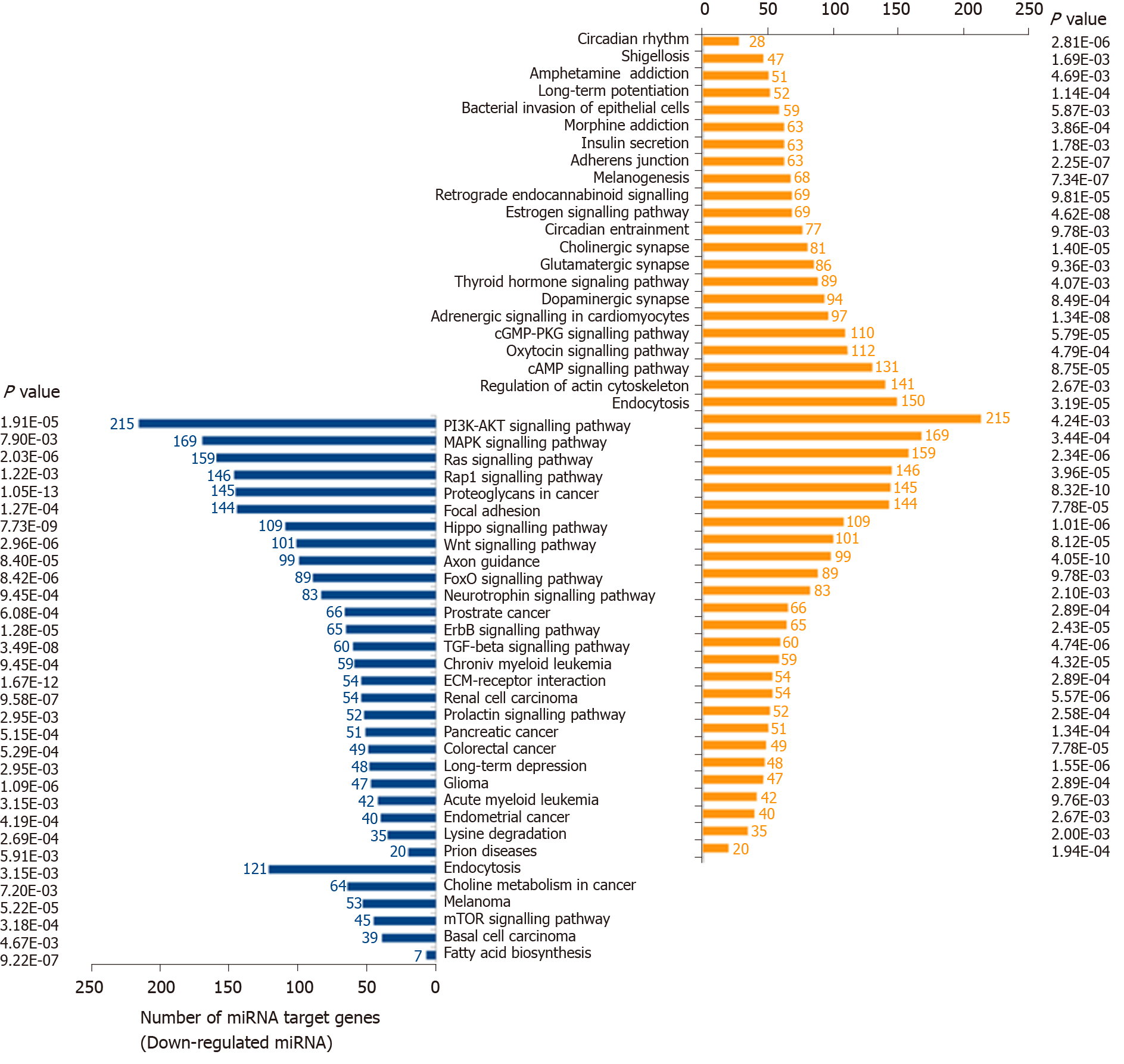
Figure 3 Pathway enrichment analyses of predicted target genes of differentially expressed microRNAs.
The differentially expressed microRNAs obtained from meta-analysis were input to DIANA miRPath version 3.0. P value thresholds were set at 0.01. PI3K: Phosphatidylinositol-3 kinases; MAPK: Mitogen-activated protein kinase.
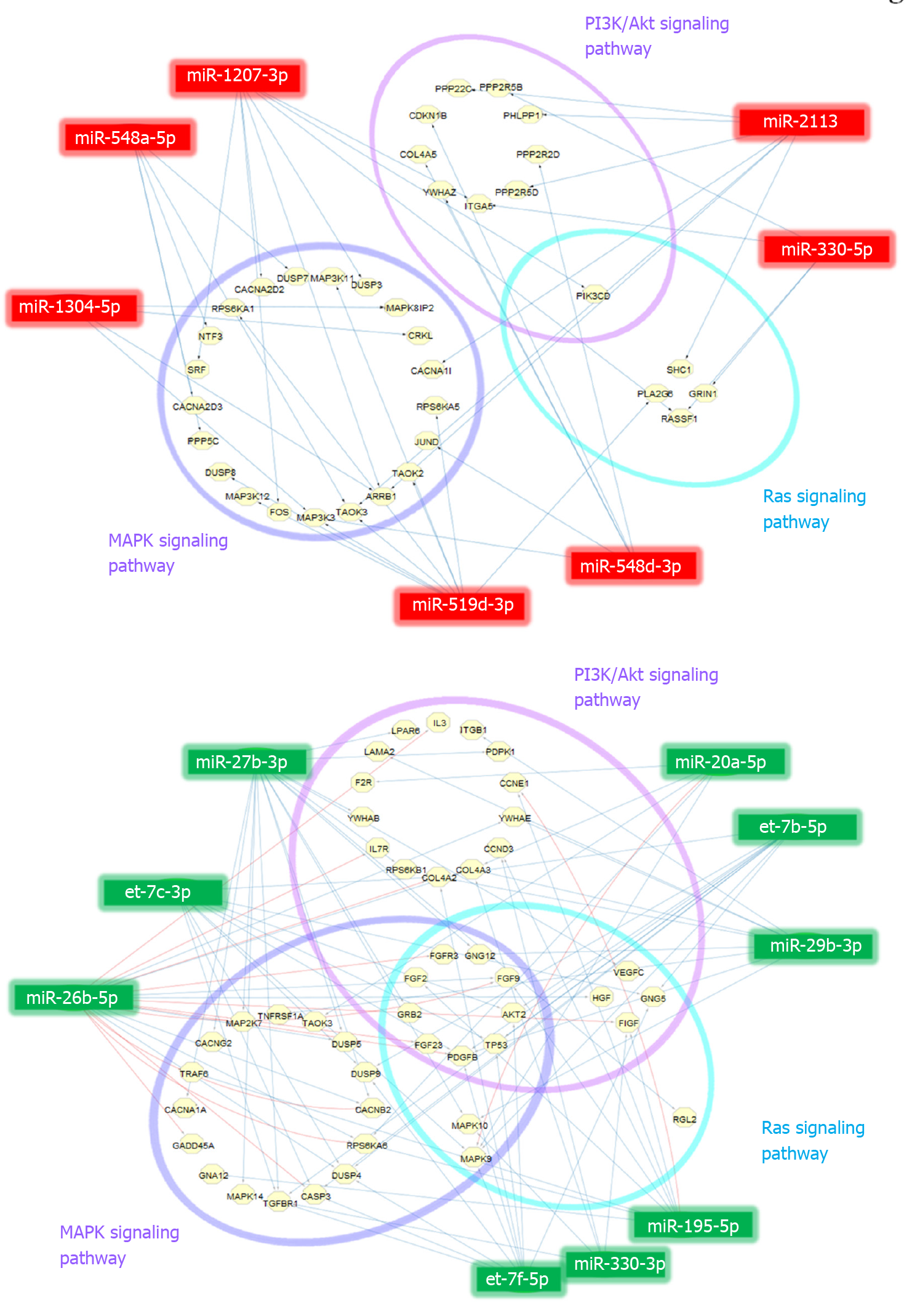
Figure 4 MicroRNA-target interaction networks.
A: Up-regulated; B: Down-regulated. MicroRNA (miRNA)-target interaction networks of 7 up-regulated and 9 down-regulated miRNAs associating in phosphatidylinositol-3 kinases/Akt, mitogen-activated protein kinase, and Ras signaling pathways based on pathway enrichment analysis via DIANA miRPath version 3.0. Blue and red lines indicate the miRNA-target prediction or information based on TargetScan and miRTarBase databases, respectively. PI3K: Phosphatidylinositol-3 kinases; MAPK: Mitogen-activated protein kinase.
- Citation: Likhitrattanapisal S, Kumkate S, Ajawatanawong P, Wongprasert K, Tohtong R, Janvilisri T. Dysregulation of microRNA in cholangiocarcinoma identified through a meta-analysis of microRNA profiling. World J Gastroenterol 2020; 26(29): 4356-4371
- URL: https://www.wjgnet.com/1007-9327/full/v26/i29/4356.htm
- DOI: https://dx.doi.org/10.3748/wjg.v26.i29.4356