Copyright
©The Author(s) 2020.
World J Gastroenterol. Aug 7, 2020; 26(29): 4272-4287
Published online Aug 7, 2020. doi: 10.3748/wjg.v26.i29.4272
Published online Aug 7, 2020. doi: 10.3748/wjg.v26.i29.4272
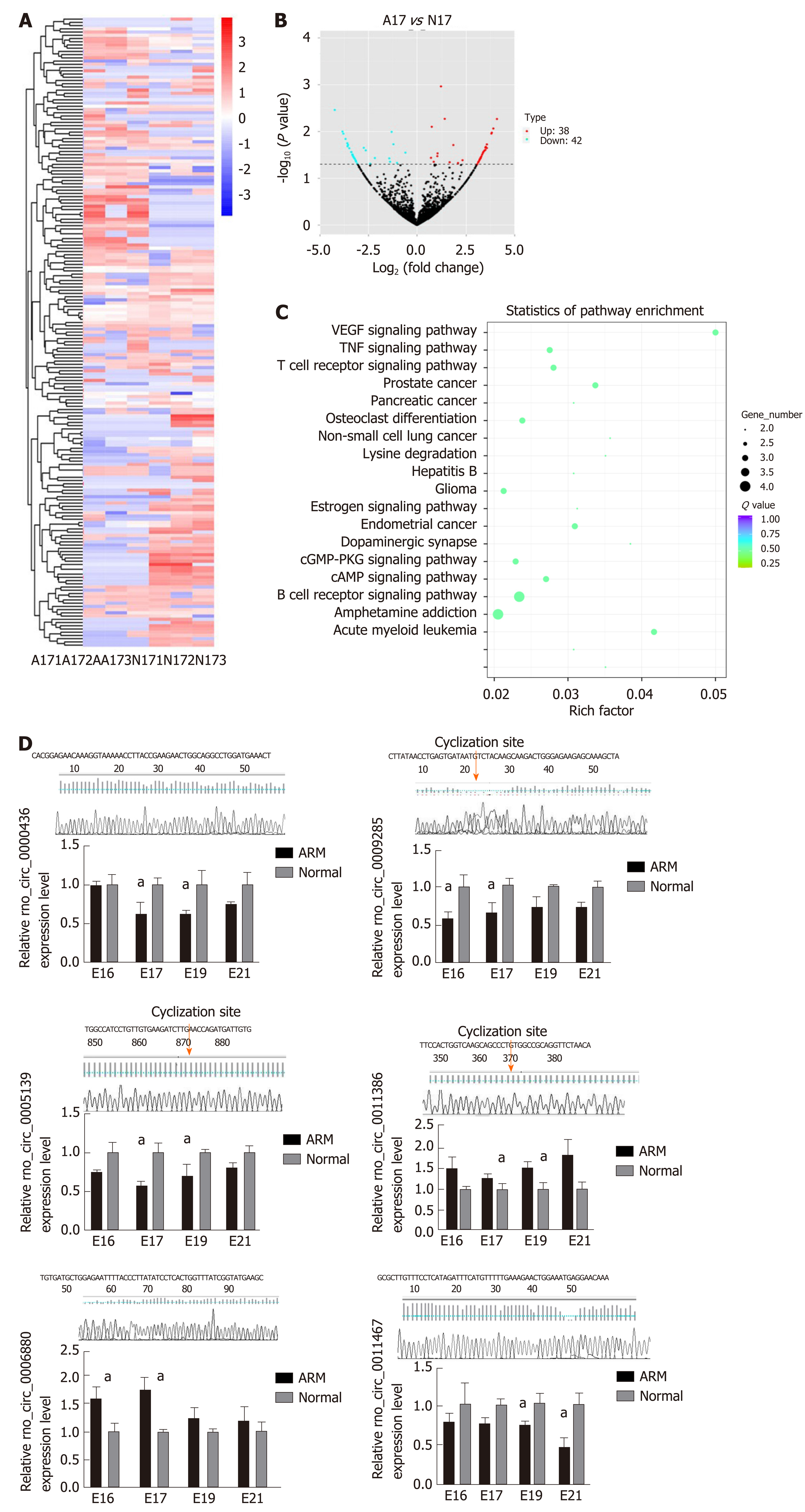
Figure 1 Identification of differentially expressed circular RNAs between anorectal malformations anorectal tissues and control anorectal tissues.
A: Cluster analysis of differentially expressed circular RNAs (circRNAs) presented as a heatmap. Red indicates high expression and blue indicates low expression [red to blue represent high to low log10 (transcripts per million + 1) values]; B: Volcano plot of 38 up-regulated (red) and 42 down-regulated (green) circRNAs on embryonic day 17 (E17) in anorectal malformations (ARM) anorectal tissues compared with control tissues showing a two-fold change; C: Vertical axis represents the pathway name, and horizontal axis represents the enrichment factor. The size of the dots indicates the number of parent genes in this pathway, and the color of the dots indicates different q-value ranges; D: circRNA rno_circ_0000436, rno_circ_0005139, rno_circ_0006880, rno_circ_0009285, rno_circ_0011386, and rno_circ_0014367 were evaluated using quantitative reverse transcription polymerase chain reaction in ARM and control anorectal tissues. aP < 0.05 vs normal. A: Anorectal malformations; ARM: Anorectal malformations; E: Embryonic day; N: Normal control.
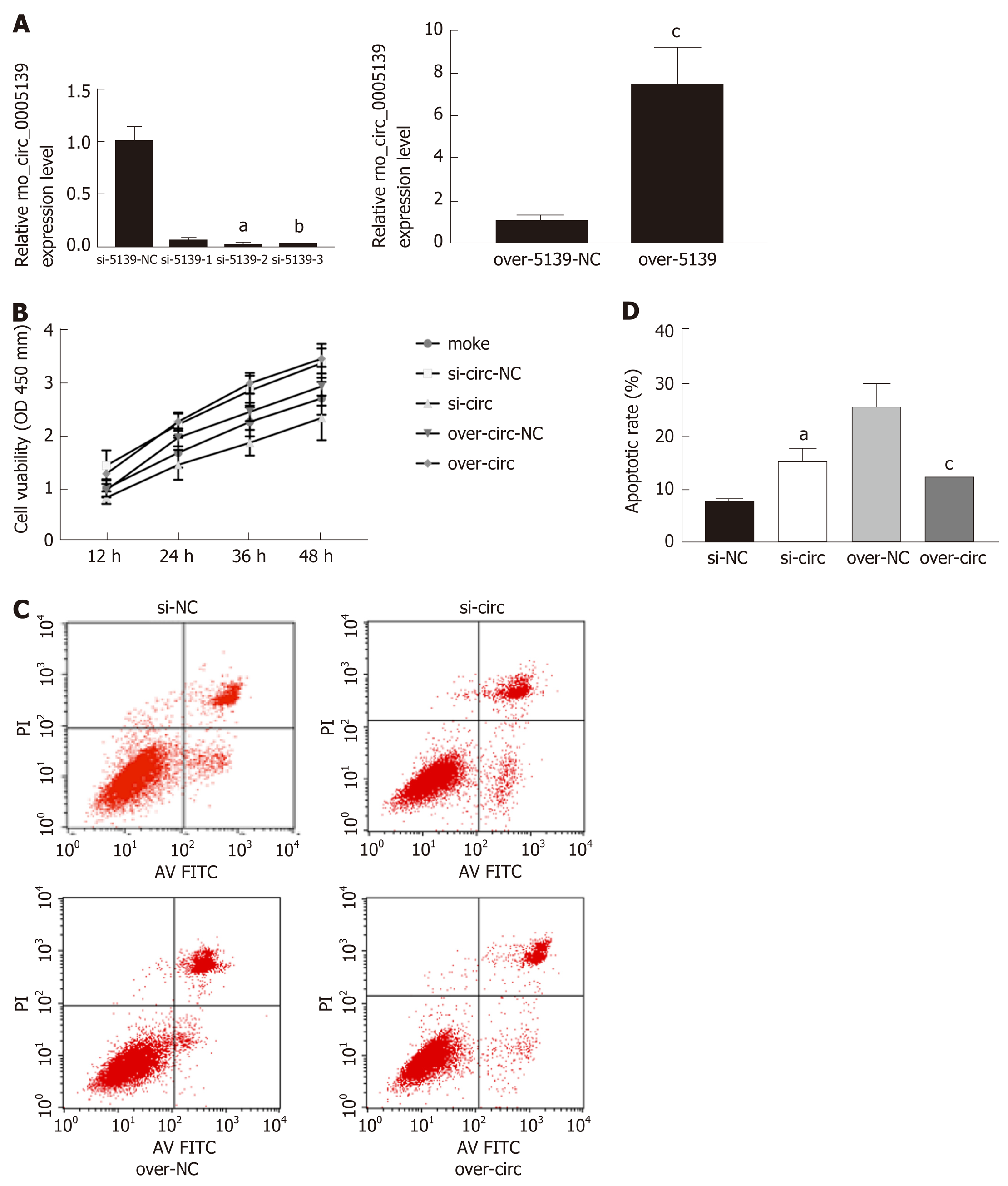
Figure 2 Knockdown and overexpression of rno_circ_0005139 affected proliferation and apoptosis of intestinal epithelial cells.
A: Expression of rno_circ_0005139 was detected after transfection with si-rno_circ_0005139 (si-circ), pLO5-rno_circ_0005139 (over-circ), or a negative control (si-NC/vector) in intestinal epithelial cells (IECs), as determined by quantitative reverse transcription polymerase chain reaction; B: Cell Counting Kit-8 assays were used to measure the viability of IECs after transfection (rno_circ_0005139); C and D: Apoptosis detection using flow cytometry was conducted to measure apoptosis in IECs after transfection (rno_circ_0005139). aP < 0.01 vs si-5139-NC, bP < 0.05 vs si-5139-NC, cP < 0.05 vs over-5139-NC.
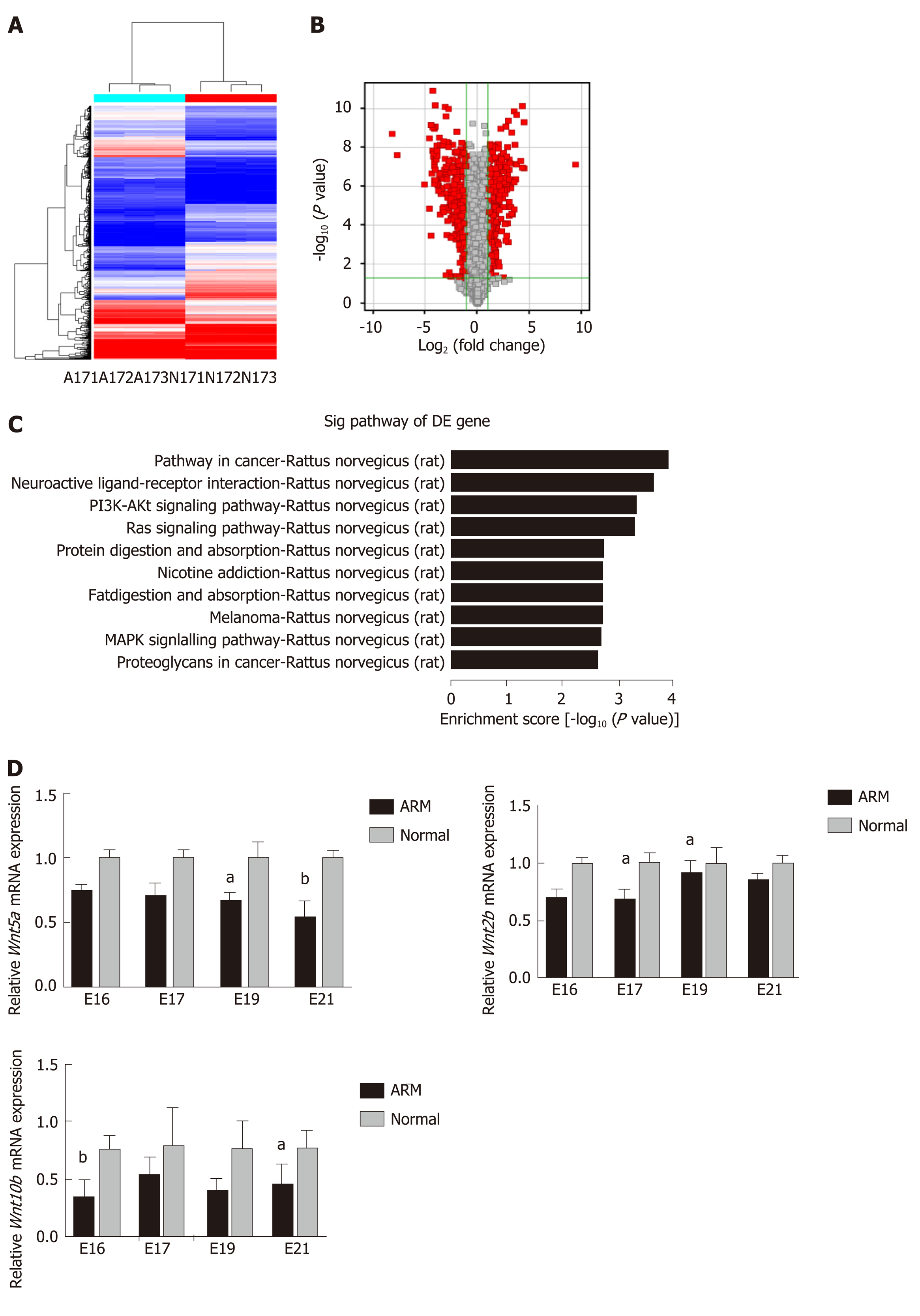
Figure 3 Identification of mRNAs in ARM and normal anorectal tissues.
A: Cluster analysis of differentially expressed mRNAs was conducted with a heatmap on E17. In the heatmap, red and blue indicate high and low expression, respectively. The color ranges from red to blue, indicating that log10 (transcripts per million + 1) expression ranges from large to small; B: Volcano plot showing 301 up-regulated (red) and 256 down-regulated (green) circular RNAs profiled on E17 in anorectal malformations (ARM) anorectal tissues compared to normal tissues showing a two-fold change (P < 0.05); C: Pathway analysis was performed using Kyoto Encyclopedia of Genes and Genomes pathways. The top 10 pathways in mRNA analysis are shown; D: Three mRNAs were evaluated by quantitative reverse transcription polymerase chain reaction in ARM and normal anorectal tissues. aP < 0.05 vs normal, bP < 0.01 vs normal. A: Anorectal malformations; ARM: Anorectal malformations; E: Embryonic day; N: Normal; DE: Differentially expression.
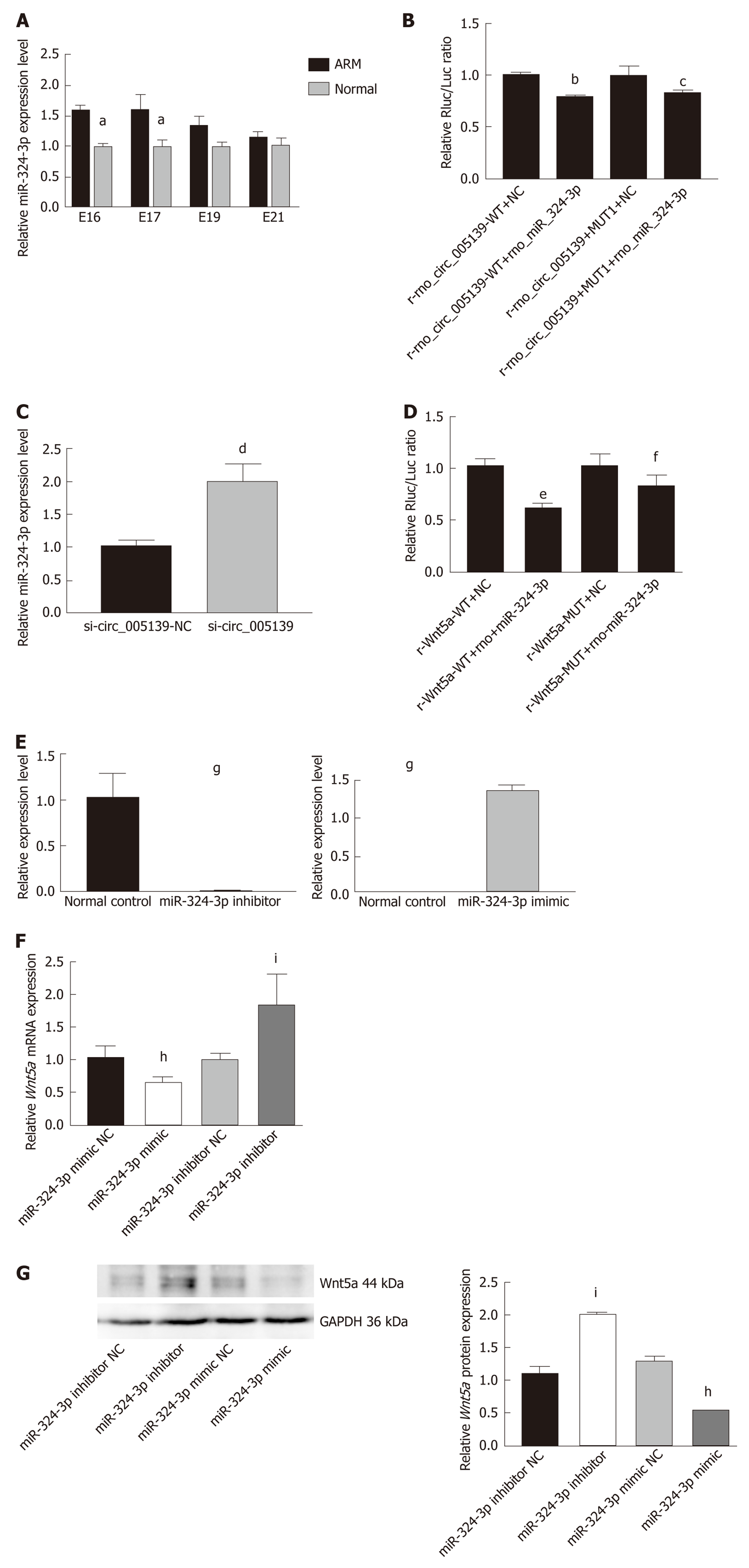
Figure 4 Rno_circ_0005139 directly binds to miR-324-3p, a direct upstream target of Wnt5a.
A: Relative expression of microRNA (miR)-324-3p in anorectal malformations and normal control anorectal tissues was detected by quantitative reverse transcription polymerase chain reaction (qRT–PCR); B: Luciferase reporter assays were conducted to explore the correlation between rno_circ_0005139 and miR-324-3p; C: Expression of miR-324-3p was detected after transfection with si-rno_circ_0005139 (si-circ) and a negative control (si-NC) in IECs by qRT–PCR; D: Luciferase reporter assays were conducted to explore the correlation between miR-324-3p and Wnt5a; E: Expression of miR-324-3p was detected after transfection with miR-324-3p mimic, miR-324-3p inhibitor, or a negative control (mimic-NC/inhibitor-NC) by qRT–PCR. Expression of Wnt5a was detected after transfection with miR-324-3p mimic, miR-324-3p inhibitor, or negative control (mimic-NC/inhibitor-NC) by qRT–PCR; F and G: mRNA and protein expression levels of Wnt5a were detected after transfection with miR-324-3p mimic, miR-324-3p inhibitor, or negative control (mimic-NC/inhibitor-NC) by qRT–PCR and western blotting. aP < 0.05 vs normal, bP < 0.01 vs r-rno_circ_0005139-WT+NC, cP < 0.05 vs r-rno_circ_0005139-MUT1+NC, dP < 0.05 vs si-circ_0005139-NC; eP < 0.05 vs r-Wnt5a-WT+NC, fP < 0.05 vs r-Wnt5a-MUT+NC, gP < 0.05 vs Normal control, hP < 0.05 vs miR-324-3p mimic NC and iP < 0.05 vs miR-324-3p inhibitor NC. ARM: Anorectal malformations; miR: microRNA.
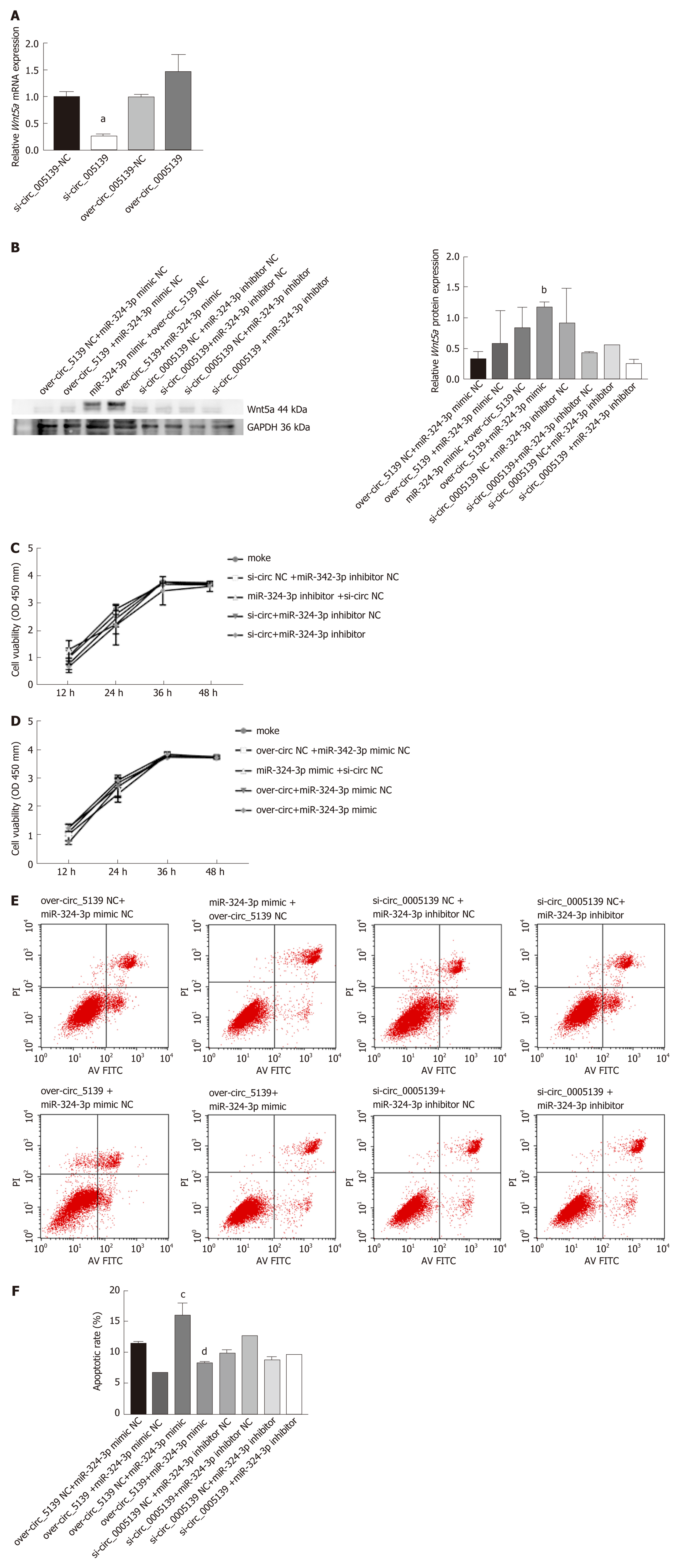
Figure 5 Rno_circ_0005139 regulated cell proliferation and apoptosis via miR-324-3p/Wnt5a.
A: Relative expression of Wnt5a was measured by quantitative reverse transcription polymerase chain reaction with si-rno_circ_0005139 (si-circ), pLO5-rno_circ_0005139 (over-circ), or a negative control (si-NC/vector); B: Wnt5a expression was measured by western blotting after co-transfection with si-rno_circ_0005139 (si-circ), miR-324-3p inhibitor, or a negative control (si-NC/inhibitor-NC), and co-transfection with pLO5-rno_circ_0005139 (over-circ), miR-324-3p mimic, or a negative control (over-NC/mimic-NC); C and D: A Cell Counting Kit-8 assay was used to measure viability in intestinal epithelial cells (IECs) after co-transfection (rno_circ_0005139 and miR-324-3p); E and F: An apoptosis assay with detection by flow cytometry was used to measure apoptosis in IECs after co-transfection (rno_circ_0005139 and miR-324-3p). aP < 0.05 vs si-circ_0005139-NC, bP < 0.05 vs over-circ_0005139-NC+miR-324-3p mimic NC, cP < 0.01 vs over-circ_0005139-NC+miR-324-3p mimic NC and dP < 0.01 vs over-circ_0005139-NC+miR-324-3p mimic.
- Citation: Liu D, Qu Y, Cao ZN, Jia HM. Rno_circ_0005139 regulates apoptosis by targeting Wnt5a in rat anorectal malformations. World J Gastroenterol 2020; 26(29): 4272-4287
- URL: https://www.wjgnet.com/1007-9327/full/v26/i29/4272.htm
- DOI: https://dx.doi.org/10.3748/wjg.v26.i29.4272