Copyright
©The Author(s) 2019.
World J Gastroenterol. Nov 14, 2019; 25(42): 6289-6298
Published online Nov 14, 2019. doi: 10.3748/wjg.v25.i42.6289
Published online Nov 14, 2019. doi: 10.3748/wjg.v25.i42.6289
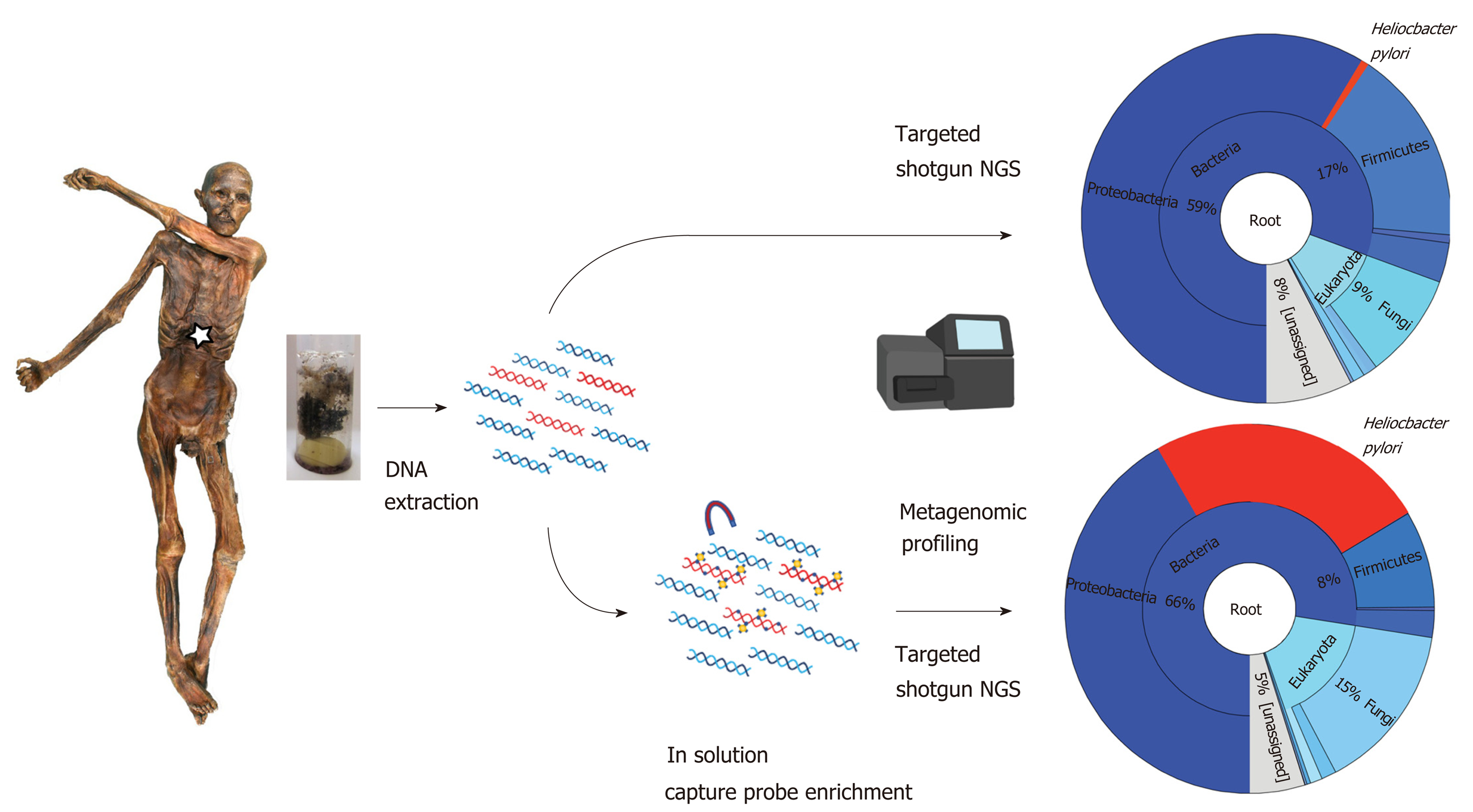
Figure 1 Molecular workflow applied to the Iceman intestinal content samples.
Both “untargeted” shotgun metagenomic next generation sequencing and “targeted” capture-enrichment have been used to identify endogenous Helicobacter pylori DNA and to reconstruct the ancient pathogen genome.
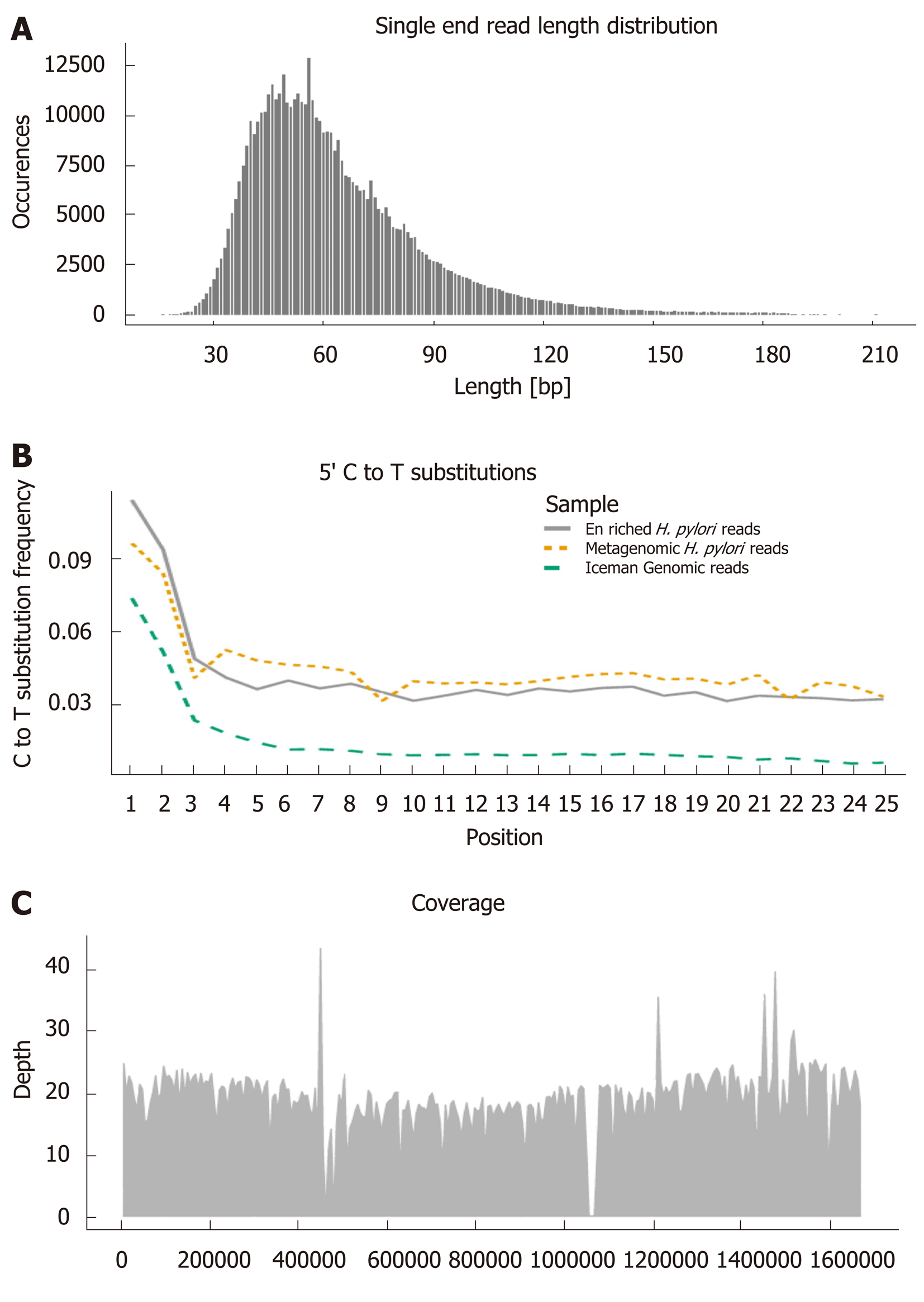
Figure 2 Ancient DNA characteristics used for authenticating endogenous sequences.
Visualised with Helicobacter pylori (H. pylori) sequences enriched from the gut of the Iceman and aligned to the H. pylori 26695 reference genome[20]: A: Read length distribution. B: C to T substitutions towards the 5’ end of sequence reads (Enriched H. pylori reads, metagenomic H. pylori reads, human Iceman genomic reads). C: Mean genome coverage and distribution of the aligned sequences. H. pylori: Helicobacter pylori.
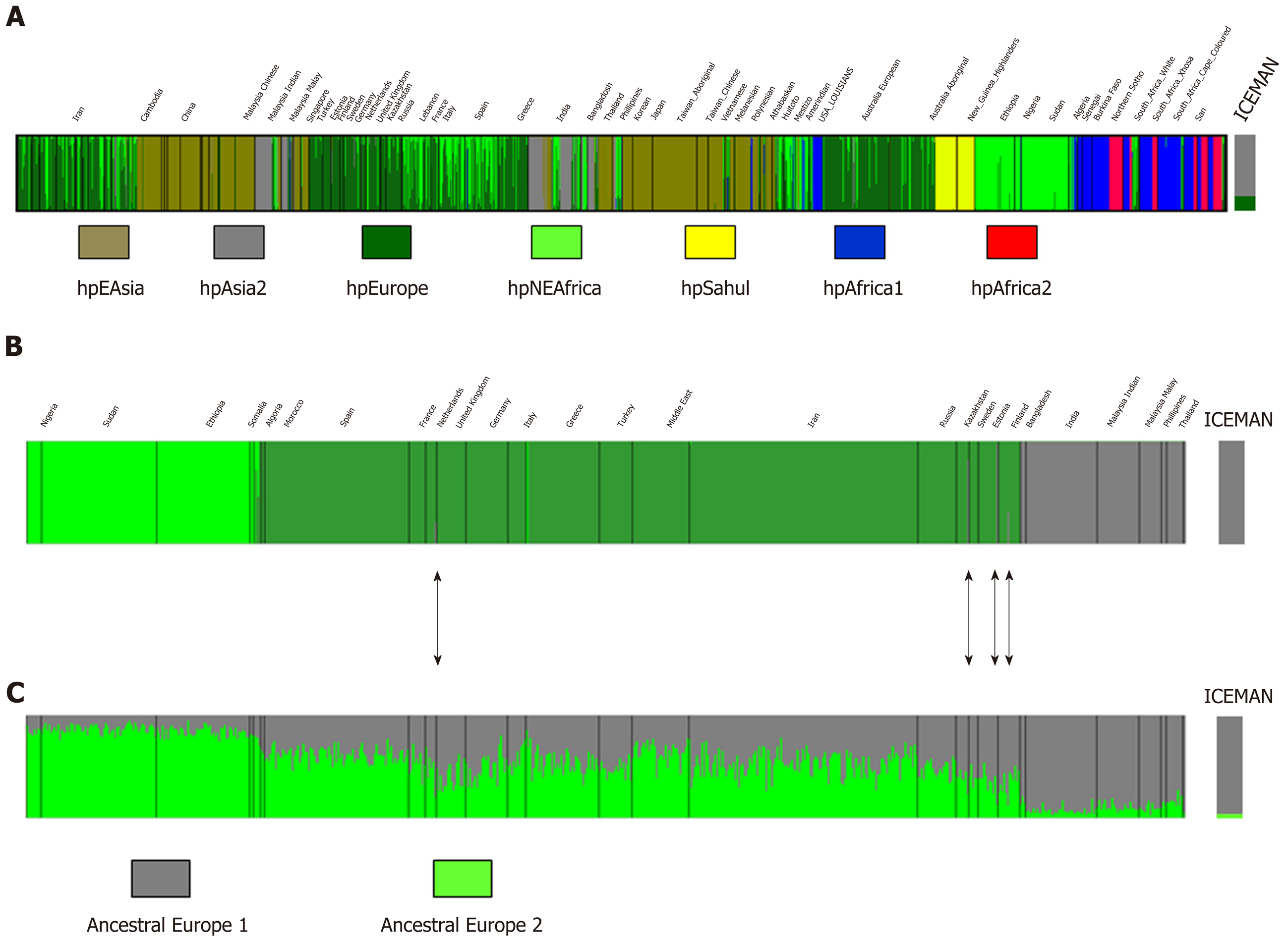
Figure 3 Population assignment of the ancient Helicobacter pylori strain using multilocus sequence typing.
A: Comparison of the Iceman strain to a collection of strains representing the worldwide Helicobacter pylori (H. pylori) populations. The proportion of the Iceman strain is displayed on the right. B: Comparison of the Iceman strain to the H. pylori populations hpNEAfrica, hpEurope, hpAsia2. C: STRUCTURE linkage model analysis showing the proportion of Ancestral Europe (AE) 1 (from Central Asia) and AE2 (from northeast Africa).
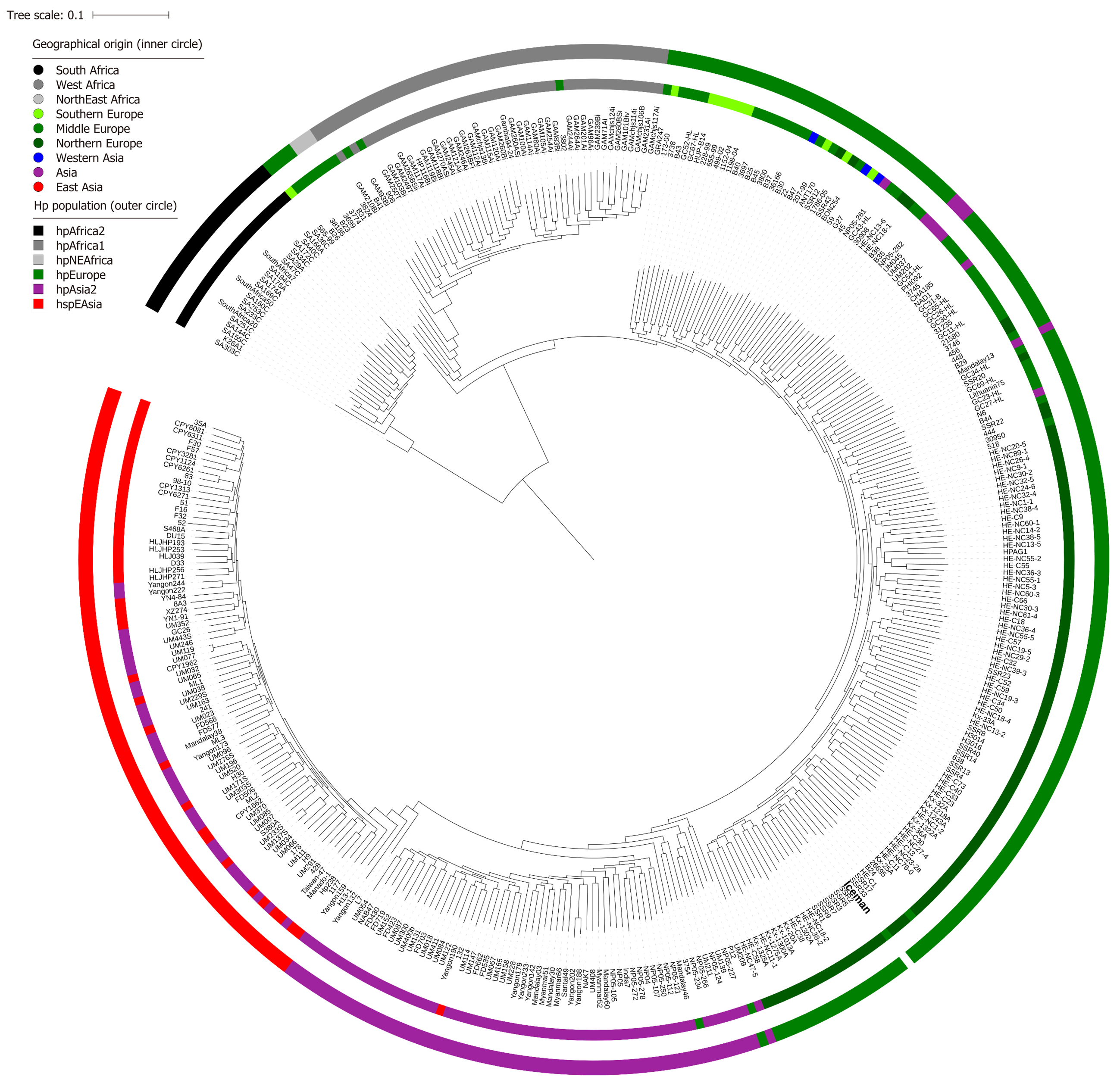
Figure 4 Helicobacter pylori core genome tree based on 1121 core genes with 375 publicly available Helicobacter pylori genomes from Europe, Asia and Africa including the Iceman (in bold).
The core genome analysis was performed using the Roary pan-genome analysis tool[44] followed by phylogenomic analysis using the PhyML tool v 3.1[45].
- Citation: Maixner F, Thorell K, Granehäll L, Linz B, Moodley Y, Rattei T, Engstrand L, Zink A. Helicobacter pylori in ancient human remains. World J Gastroenterol 2019; 25(42): 6289-6298
- URL: https://www.wjgnet.com/1007-9327/full/v25/i42/6289.htm
- DOI: https://dx.doi.org/10.3748/wjg.v25.i42.6289