Copyright
©The Author(s) 2019.
World J Gastroenterol. Sep 7, 2019; 25(33): 4985-4998
Published online Sep 7, 2019. doi: 10.3748/wjg.v25.i33.4985
Published online Sep 7, 2019. doi: 10.3748/wjg.v25.i33.4985
Figure 1 Primer and LNA-probe positions designed for the detection of hepatitis B virus rtM204I (YIDD) variant and rtM204 (YMDD) wild type.
Arrows indicate the primer positions. Underlines indicate the probe positions. The numbers designate the nucleotide position on the hepatitis B virus reverse transcriptase gene sequence. Boldface bases denote the different bases. The box represents the codon and amino acid sequences of the rtM204 (YMDD) wild type and rtM204I (YIDD) variant. This single nucleotide difference is the basis of their discriminative identification by LNA probes in this study. The amino acid sequence is shown as the one-letter amino acid symbols. WT: Wild type.
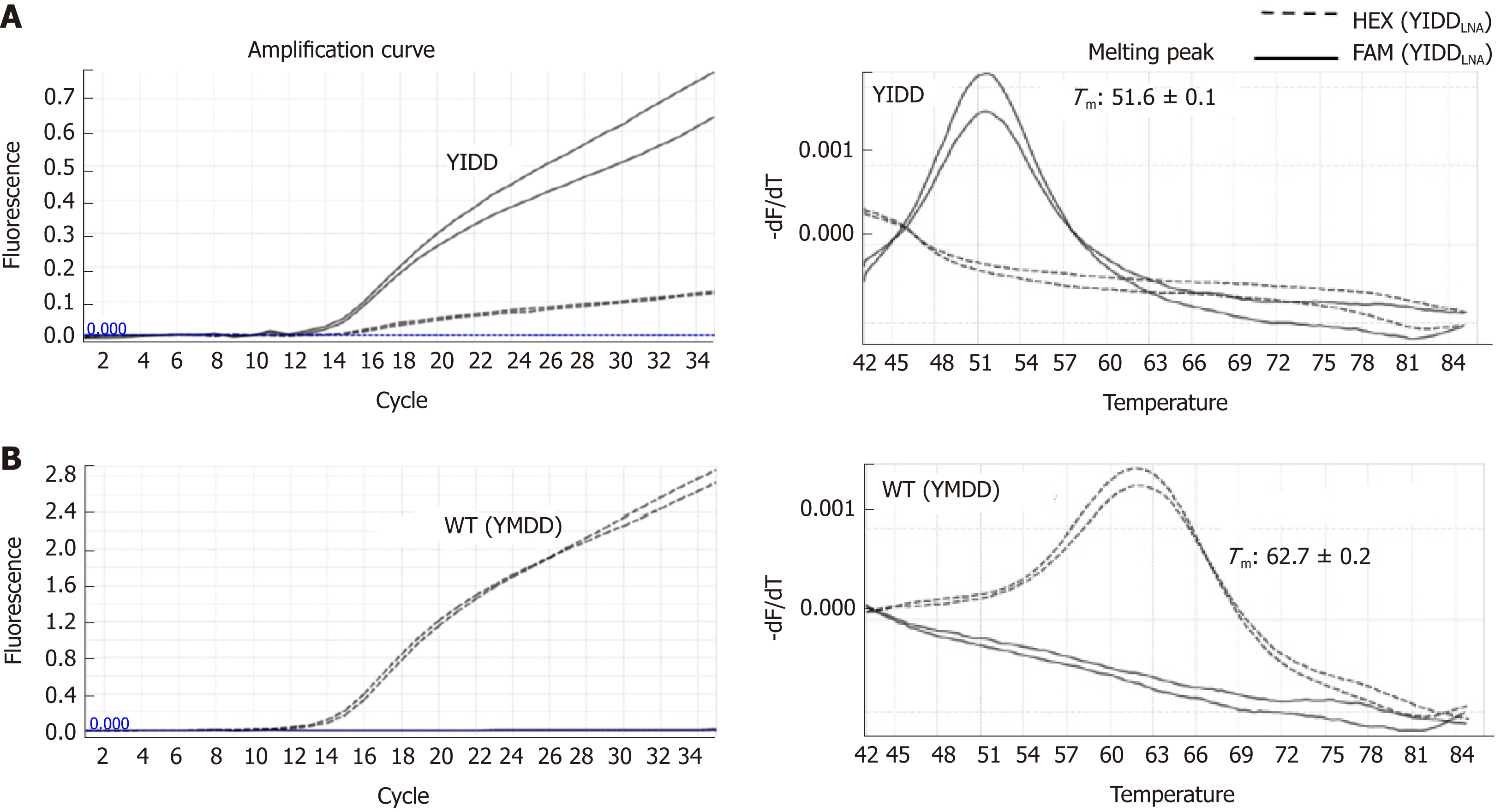
Figure 2 LNA real-time PCR for identification of hepatitis B virus rtM204I (YIDD) variant and rtM204 (YMDD) wild type.
Amplification curves were shown on the left, melting peaks on the right. With YIDD variant DNA templates (A), YIDD specific signals at FAM channel (solid) were detected showing their dominant amplifications with minimal cross signals of amplification generated by a weak YMDD probe cross hybridization and distinct melting temperatures different from those of YMDD DNA with no significant cross signals on melting. For WT DNA templates (B), YMDD specific signals at HEX 6 channel (dashed) were detected showing their exclusive amplifications and distinct Tms different from those of YIDD with no cross signals. WT: Wild type.
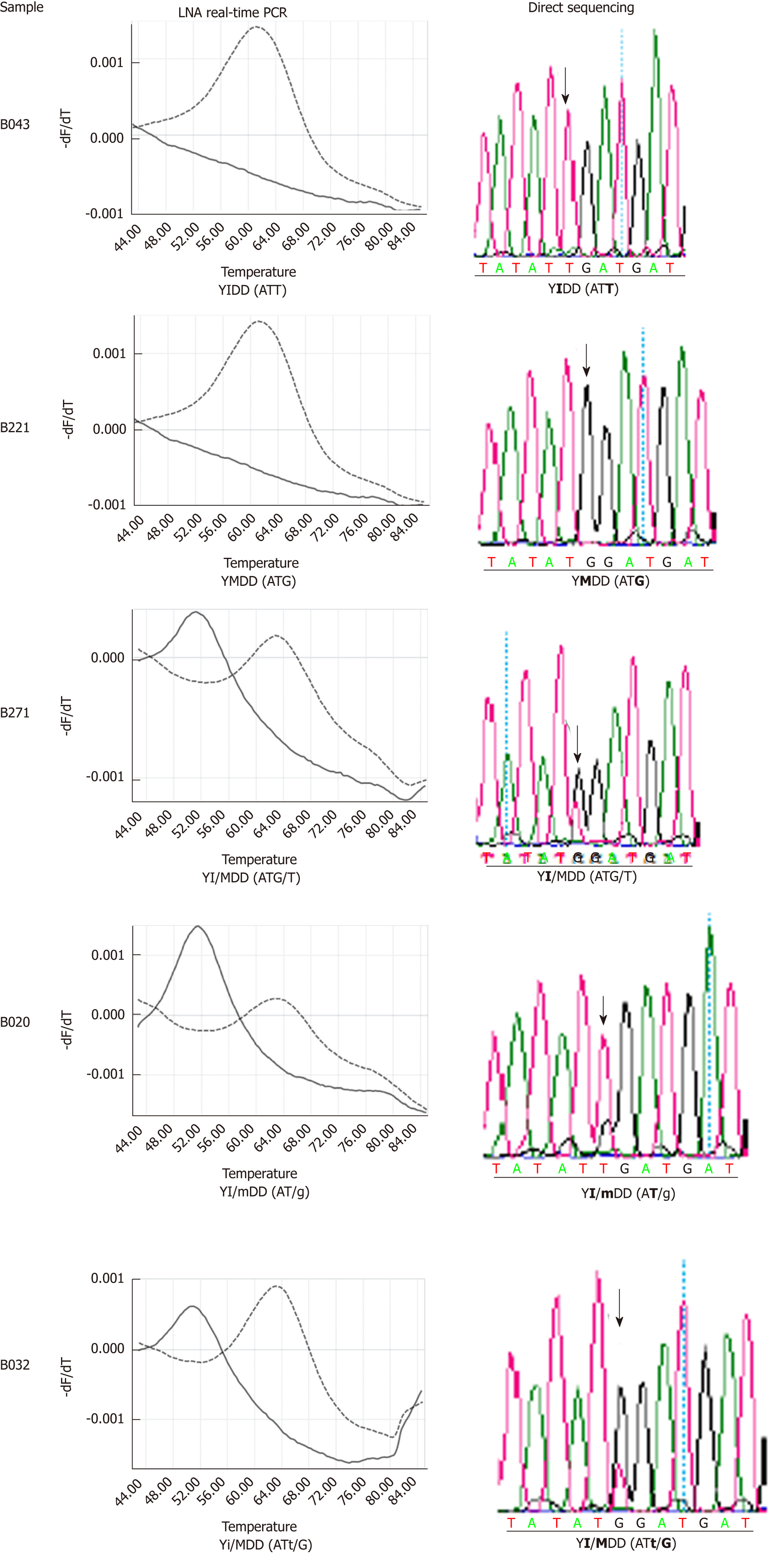
Figure 3 Confirmation of LNA real-time PCR identification results of hepatitis B virus rtM204I (YIDD) variant and rtM204 (YMDD) wild type by direct sequencing.
Nucleotide bases are shown in the parenthesis. Lower case letters represent the base in a lower amount relative to the dominant variant. Bold indicates the target amino acid and bases.
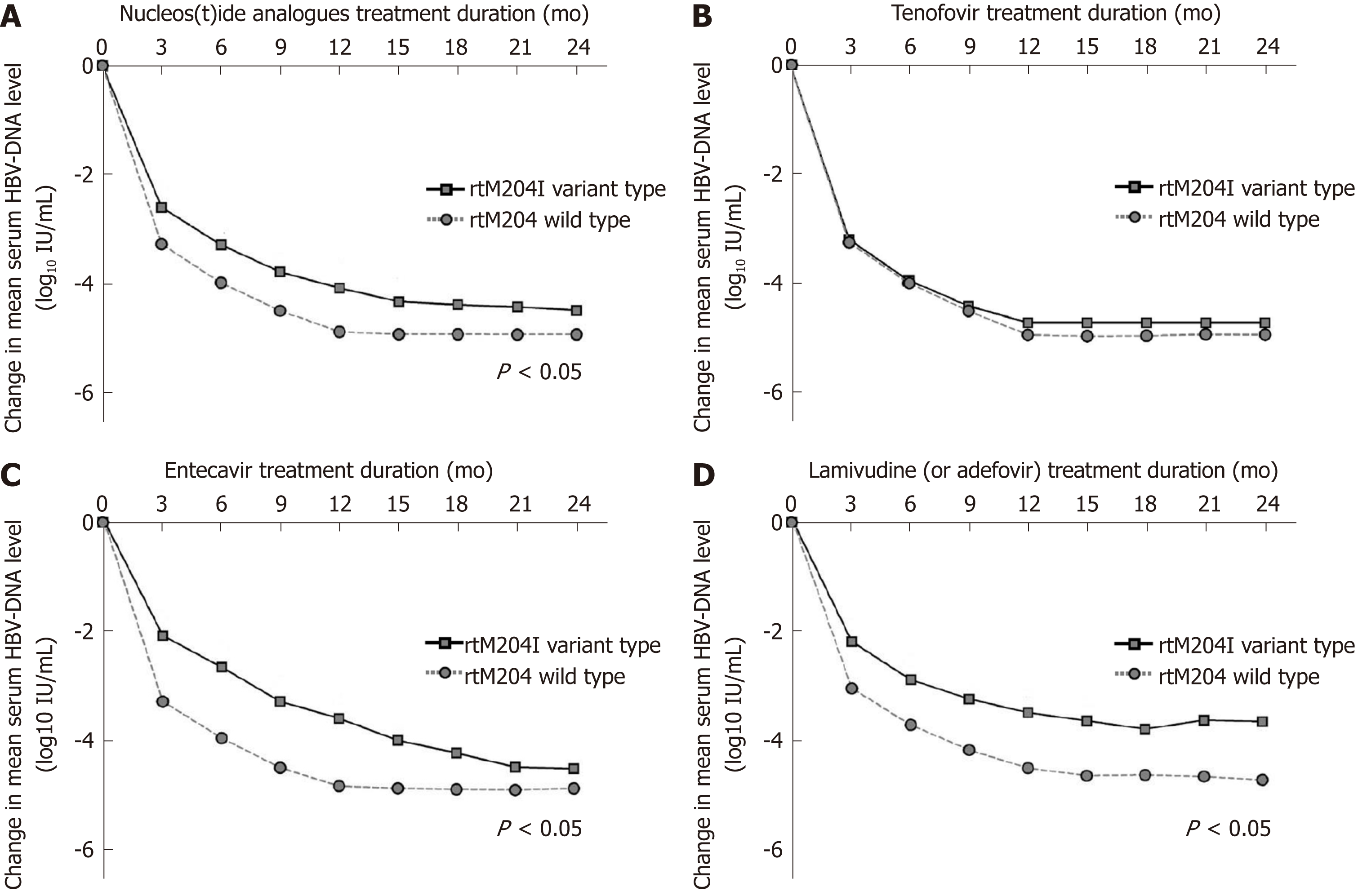
Figure 4 Changes in mean log values of the serum hepatitis B virus DNA levels from baseline during nucleos(t)ide analogues treatment.
A: The decrease in hepatitis B virus (HBV) DNA was significantly less prominent in patients infected with naturally occurring rtM204I variants than in patients without pre-existing rtM204I variants at 3, 6, 9, 12, 15 mo of nucleos(t)ide analogues (all P < 0.05); B: There was no differences in HBV-DNA declines during tenofovir therapy between patients with and without naturally occurring rtM204I variants; C: The decrease in HBV DNA was significantly less prominent in patients infected with naturally occurring rtM204I variants than in patients without pre-existing rtM204I variants at 3, 6, 9, 12, 15 mo of entecavir (all P < 0.05); D: The decrease in HBV DNA was significantly less prominent in patients infected with naturally occurring rtM204I variants than in patients without pre-existing rtM204I variants at 3, 6, 9, 12, 15 mo of low genetic barriers (all P < 0.05). Student's t-test was used for the statistical analysis at each time point. HBV: Hepatitis B virus.
- Citation: Choe WH, Kim K, Lee SY, Choi YM, Kwon SY, Kim JH, Kim BJ. Tenofovir is a more suitable treatment than entecavir for chronic hepatitis B patients carrying naturally occurring rtM204I mutations. World J Gastroenterol 2019; 25(33): 4985-4998
- URL: https://www.wjgnet.com/1007-9327/full/v25/i33/4985.htm
- DOI: https://dx.doi.org/10.3748/wjg.v25.i33.4985