Copyright
©The Author(s) 2019.
World J Gastroenterol. Aug 28, 2019; 25(32): 4661-4672
Published online Aug 28, 2019. doi: 10.3748/wjg.v25.i32.4661
Published online Aug 28, 2019. doi: 10.3748/wjg.v25.i32.4661
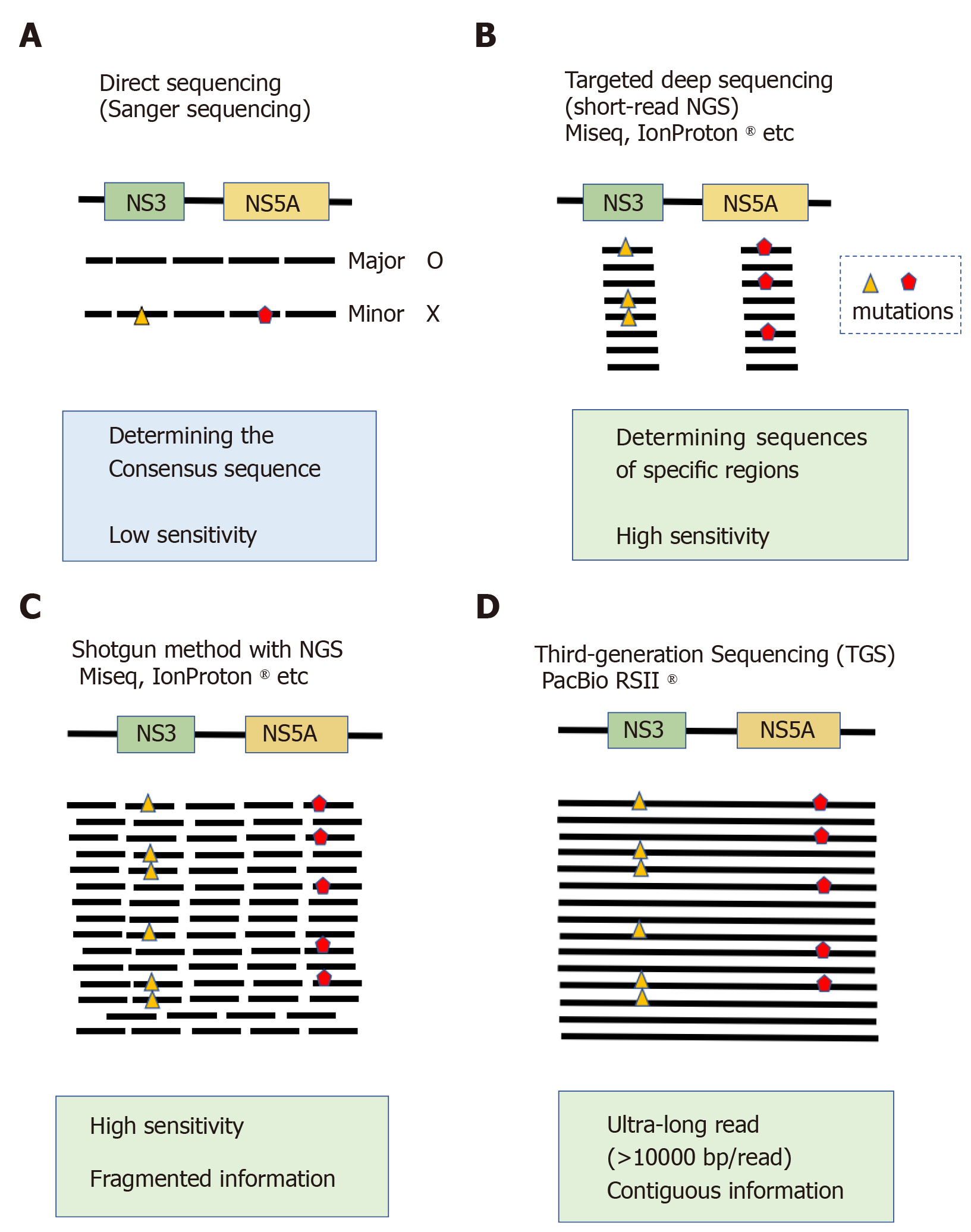
Figure 1 Comparison of sequencing platforms.
A: Direct sequencing (Sanger sequencing). This conventional sequencing method determines the consensus sequence of target regions. Nucleotide variants with allele frequencies of approximately 15% can be detected; B: Targeted deep sequencing using conventional short-read next-generation sequencing (NGS) can detect low abundance variants making up approximately 1% of total mapped reads; C: When long PCR products are used as templates for conventional short-read NGS, they are first fragmented into 100-200 bp segments, ligated to sequence adapters, amplified and then sequenced. The sequenced reads are mapped to a reference sequence using the shotgun method. One of the limitations of this technique is a lack of information regarding whether two distant mutations co-exist on a single template molecule; D: Third-generation sequencing methods represented by single-molecular real-time sequencing can generate ultra-long reads of more than 10000 bp, and contiguous sequence information can be obtained. NGS: Next-generation sequencing.
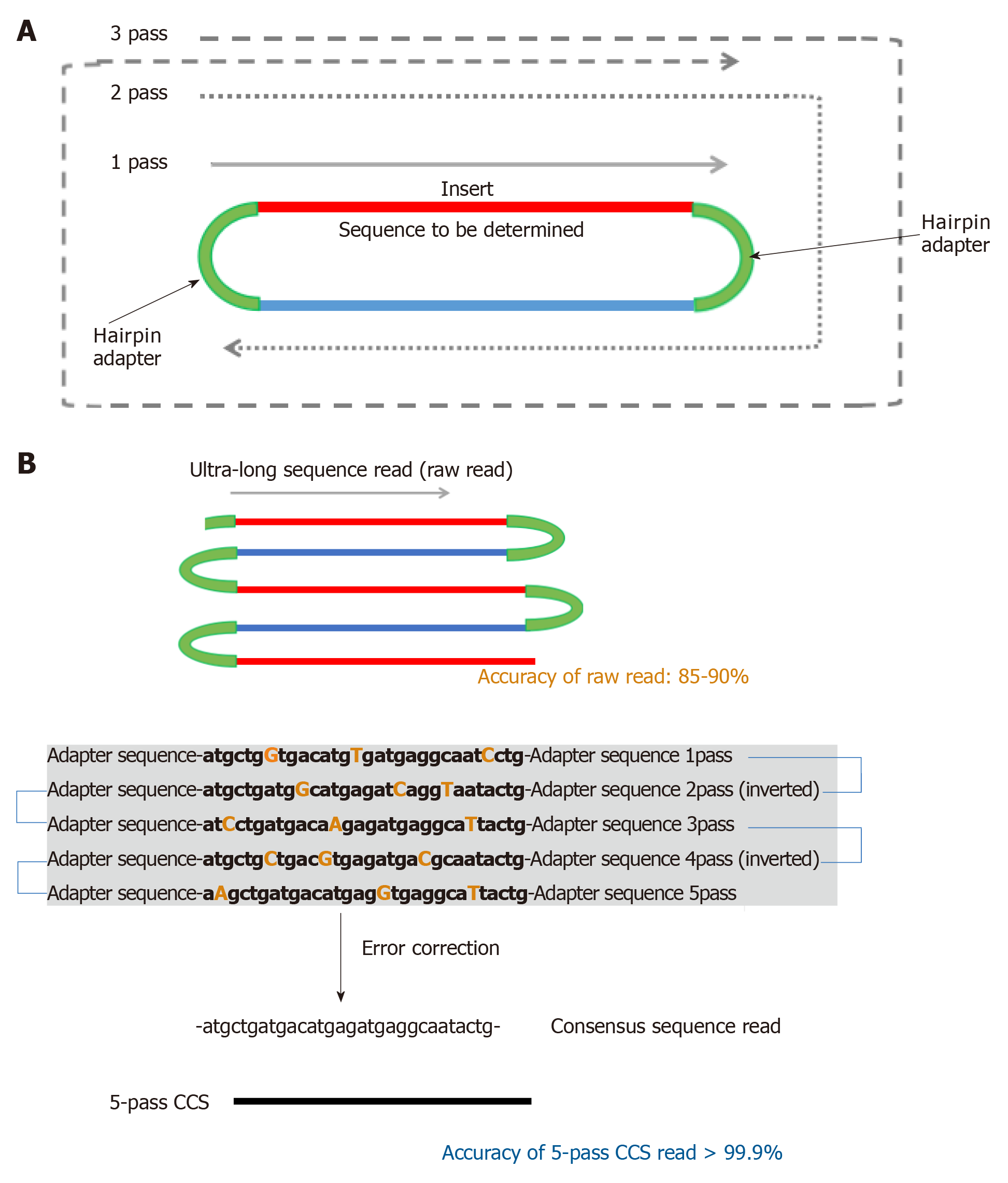
Figure 2 Generation of circular consensus sequences.
A: The template for PacBio sequencing, called SMRTBell, is created by ligating hairpin adaptors to both ends of a double-stranded DNA molecule containing the sequence to be determined. This template then acts like a single-stranded closed circle. The polymerase initiates at the primer location and sequences the template until it falls off. The enzyme then proceeeds around the hairpin on the other end of the SMRTBell, and can circle around the same template multiple times; B: Scheme for generation of 5-pass circular consensus sequences (CCS) reads. Ultra-long raw reads are generated by a polymerase. Although the accuracy of the raw read is 85%-90%, error-corrected consensus reads (CCS reads) can be generated using the data from a single template sequenced multiple times. The accuracy of 5-pass CCS reads is as high as 99.9%. CCS: Circular consensus sequences.
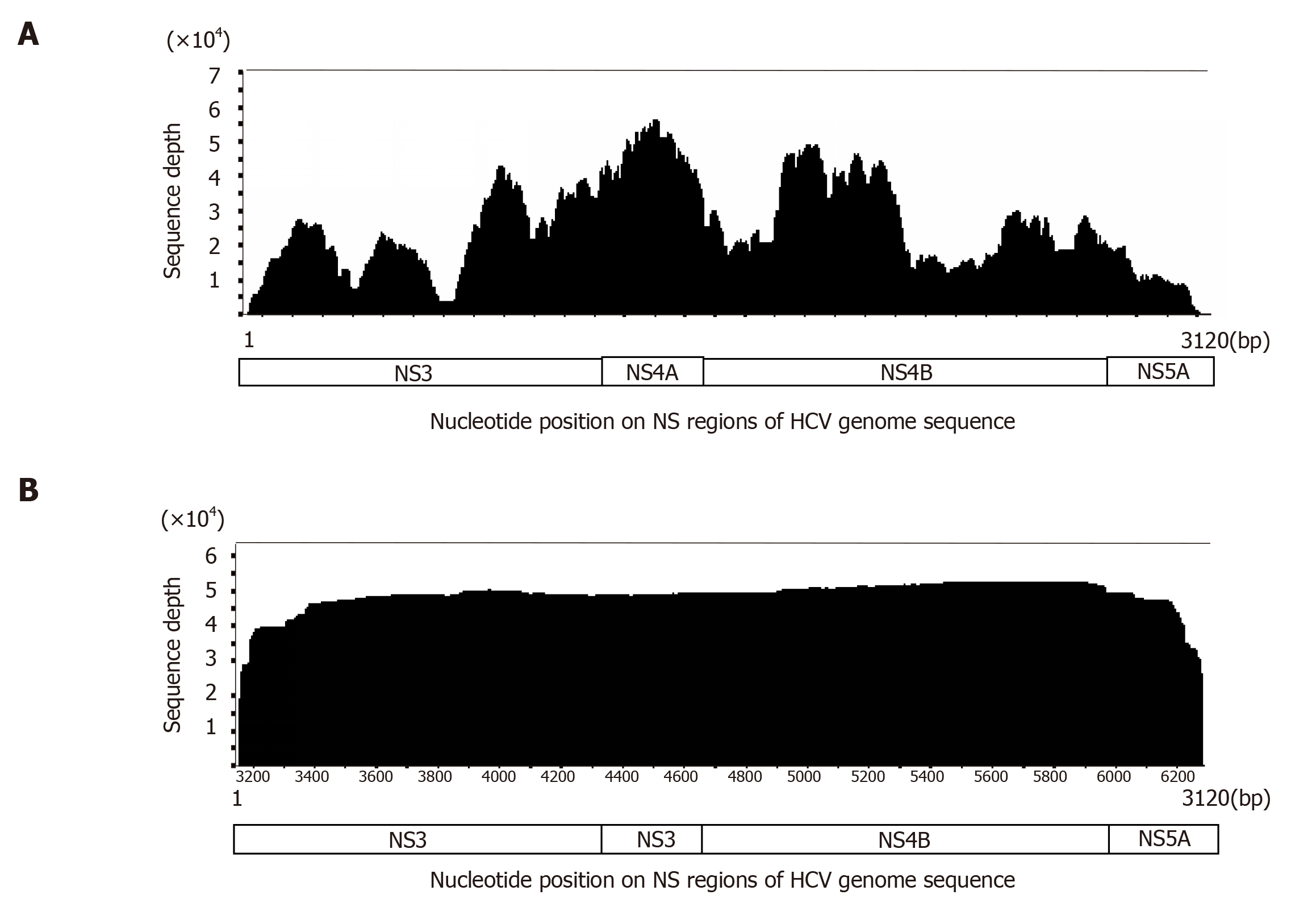
Figure 3 Comparison of coverage curves generated by short-read next-generation sequencing and long-read single-molecular real-time sequencing.
A: A coverage curve generated by an IonProton sequencer. Approximately 3120 bp from the NS3 to NS5A region of the hepatitis C virus (HCV) genome from an HCV-infected patient was amplified and long-PCR products were subjected to short-read sequencing. The sequencing depth varies according to genomic location; B: When the same template was sequenced using PacBio RSII sequencer, the coverage curve demonstrates uniform coverage through the NS3 to NS5A regions. HCV: Hepatitis C virus.
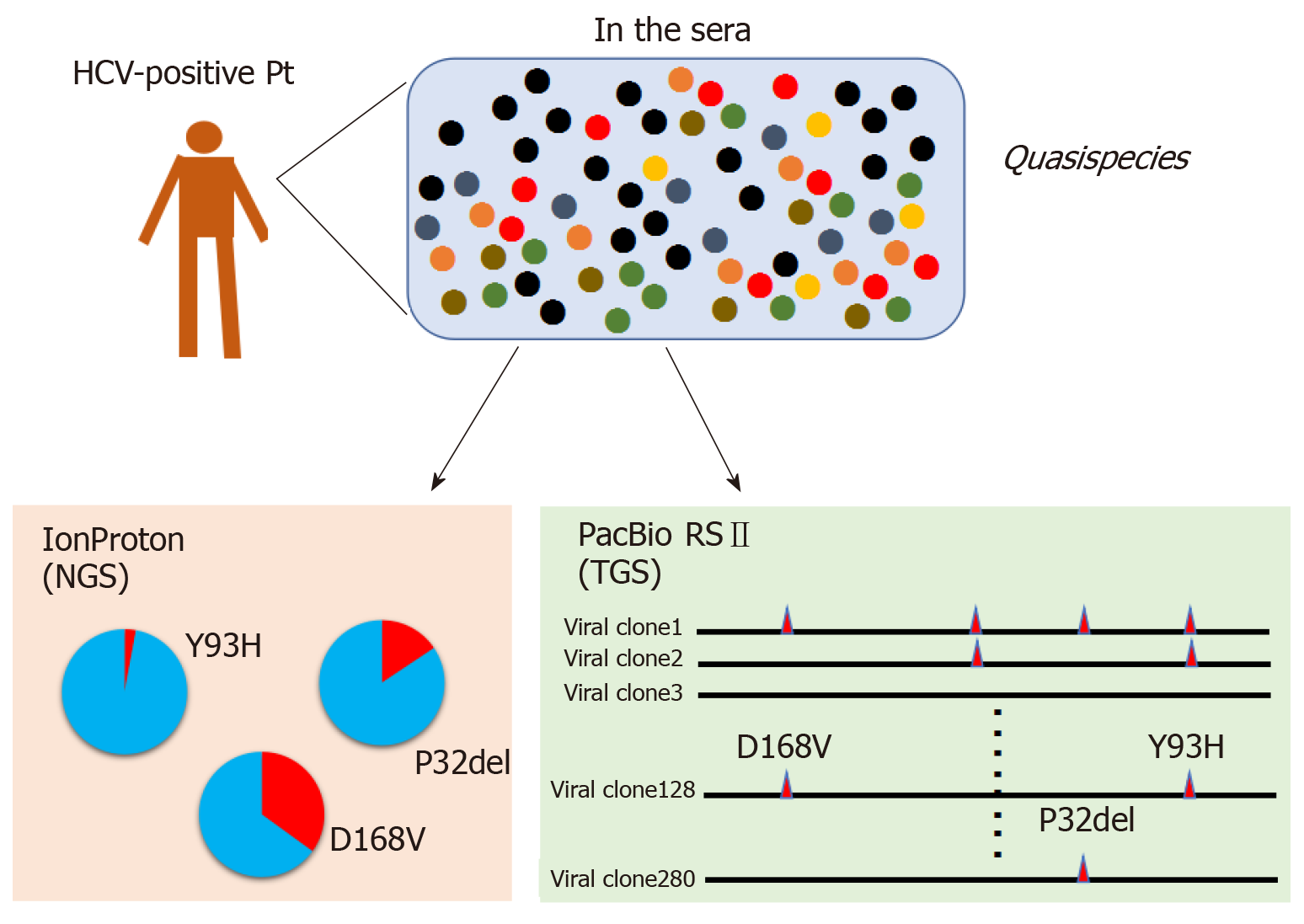
Figure 4 Comparison of short-read and long-read sequencing for analysis of viral quasispecies.
Conventional short-read sequencing, such as the IonProton sequencer, generates bulk information on viral clones. However, only fragmented information can be obtained such as the frequency of viral clones bearing the NS3-D168V or NS5A-P32del variants. By contrast, PacBio RSII sequencing can determine the contiguous genome sequence of each template, permitting analysis of linkage between several nucleotide changes through the NS3 to NS5A regions for individual viral clones. TGS: Third-generation sequencing; NGS: Next-generation sequencing; HCV: Hepatitis C virus.
- Citation: Takeda H, Yamashita T, Ueda Y, Sekine A. Exploring the hepatitis C virus genome using single molecule real-time sequencing. World J Gastroenterol 2019; 25(32): 4661-4672
- URL: https://www.wjgnet.com/1007-9327/full/v25/i32/4661.htm
- DOI: https://dx.doi.org/10.3748/wjg.v25.i32.4661