Copyright
©The Author(s) 2019.
World J Gastroenterol. May 28, 2019; 25(20): 2489-2502
Published online May 28, 2019. doi: 10.3748/wjg.v25.i20.2489
Published online May 28, 2019. doi: 10.3748/wjg.v25.i20.2489
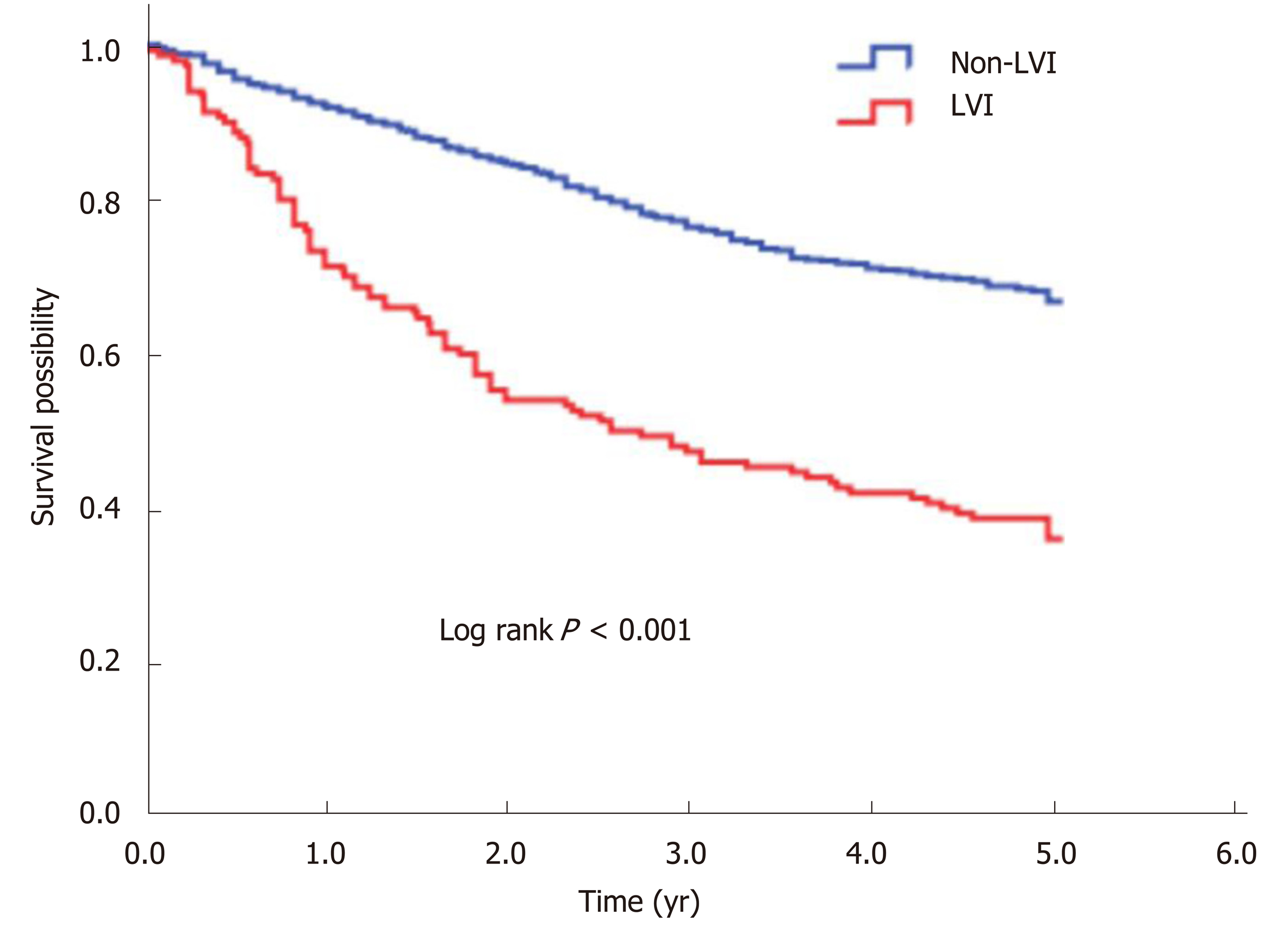
Figure 1 Kaplan-Meier curve for 5-year overall survival according to LVI status.
The 5-year overall survival rate of LVI group was significantly lower than that of non-LVI group (36.0% vs 66.8%, log-rank P < 0.001). LVI: Lymphovascular invasion.
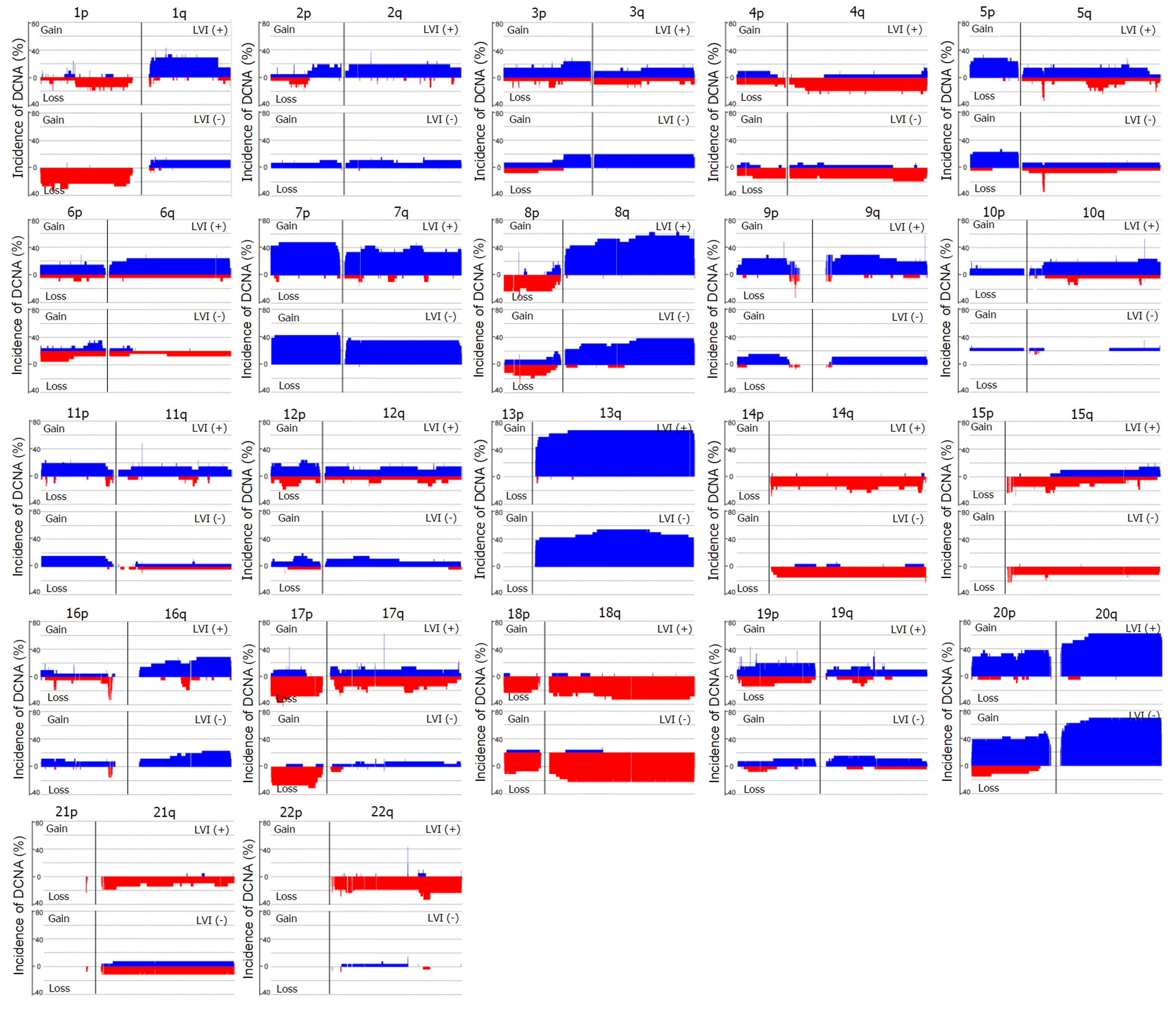
Figure 2 Comparison of DNA copy number alterations between colorectal cancers with and without LVI.
Data were shown for the autosomes only. Blue represents gain of expression; red represents loss of expression. One hundred eighty-four DNA copy number alterations (105 gains and 79 losses) exhibited a significant difference in their frequencies between the two groups (P < 0.05). DCNA: DNA copy number alteration; LVI: Lymphovascular invasion.
Figure 3 DCNA classifier for differentiating between colorectal cancers with and without LVI.
LVI were distinguished from those without it at a 95.7% accuracy rate by examining seven special DCNAs. The procedures of this analysis were as follows: chr6: 95148725-95193920 was taken as the first criterion. If a tumor showed gain at chr6: 95148725-95193920, it was categorized as a tumor with LVI. Six of 47 tumors were classified into this group. When the DCNA of the tumor was not this case, the second criterion (Loss at chr17: 30770711-30859316) was examined. Five tumors were separated from the remaining 41 tumors at this point. If the tumors showed no loss at chr17: 30770711-30859316, then the third criterion (Gain at chr14: 106113380-106135655) was checked. In this way, the DCNA of the tumor was in turn examined from the 1st to the 7th one. Each step successively sorted a cluster of either group. Finally, all colorectal cancers were divided into either of two groups, tumors with and without LVI. DCNA: DNA copy number alteration; LVI: Lymphovascular invasion.
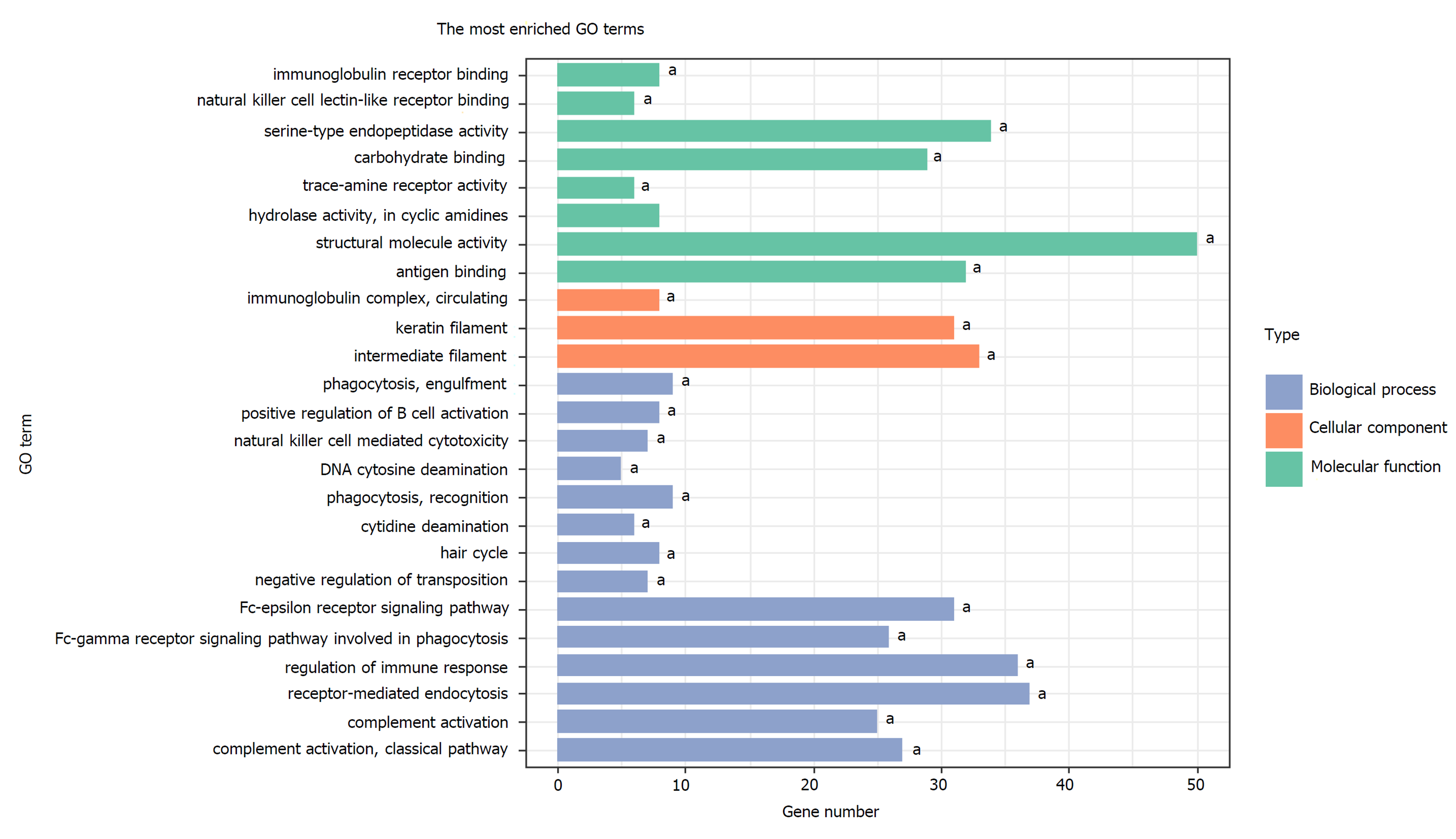
Figure 4 Top GO terms enriched by the 1784 genes located at the 184 significant DNA copy number alterations.
aP < 0.05; GO: Gene ontology.
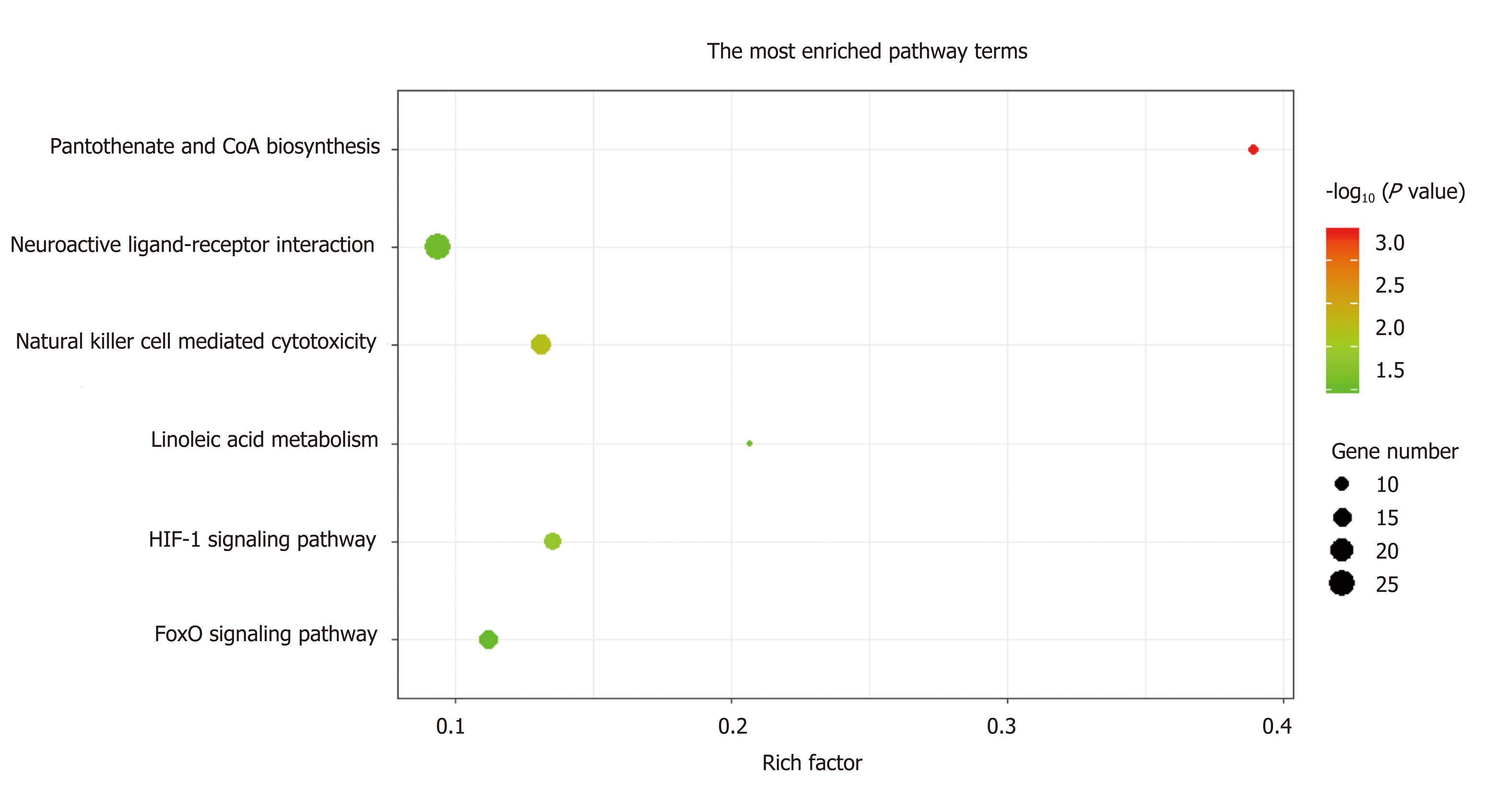
Figure 5 Top Kyoto Encyclopedia of Genes and Genomes pathway terms enriched by the 1784 genes located at the 184 significant DNA copy number alterations.
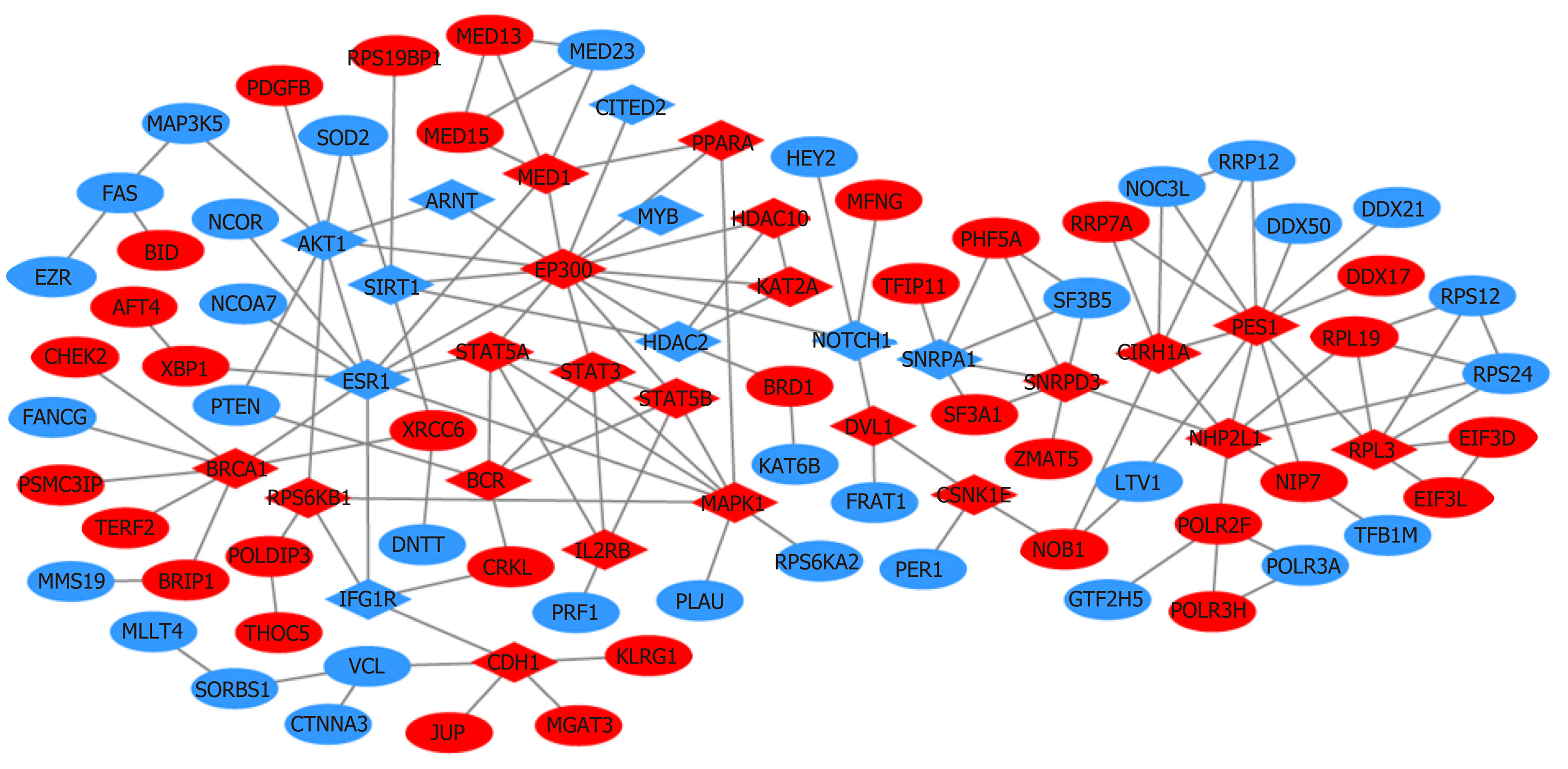
Figure 6 Protein‑protein interaction network for the genes related to lymphovascular invasion (confidence score > 0.
95). Blue represents gain of expression; red represents loss of expression. Diamonds represent the 31 hug genes identified by closeness, degree, and eigenvector.
- Citation: Jiang HH, Zhang ZY, Wang XY, Tang X, Liu HL, Wang AL, Li HG, Tang EJ, Lin MB. Prognostic significance of lymphovascular invasion in colorectal cancer and its association with genomic alterations. World J Gastroenterol 2019; 25(20): 2489-2502
- URL: https://www.wjgnet.com/1007-9327/full/v25/i20/2489.htm
- DOI: https://dx.doi.org/10.3748/wjg.v25.i20.2489