Copyright
©The Author(s) 2019.
World J Gastroenterol. Apr 21, 2019; 25(15): 1783-1796
Published online Apr 21, 2019. doi: 10.3748/wjg.v25.i15.1783
Published online Apr 21, 2019. doi: 10.3748/wjg.v25.i15.1783
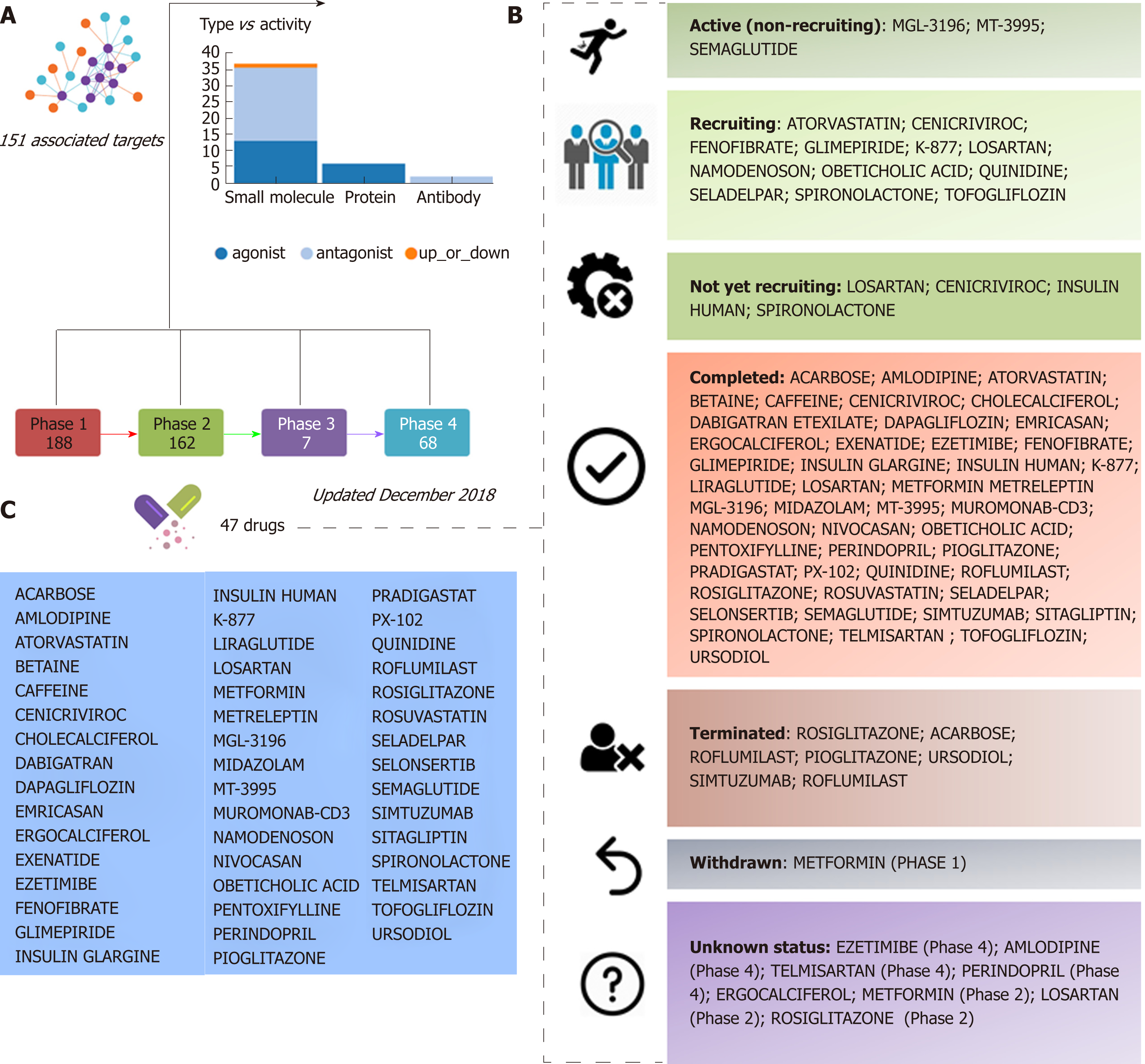
Figure 1 Clinical trials for the treatment of nonalcoholic steatohepatitis.
A and B: Figure highlights 47 drugs that are currently under investigation for the treatment of nonalcoholic steatohepatitis in different pharmacological phases (from phase 1 to phase 4): Information on clinical trial status (recruitment status) as well as prediction of potential associated targets were retrieved from the Target Validation Platform available at https://http://www.targetvalidation.org; C: Drugs listed in the most advanced pharmacological phase updated December 2018 concerning to privately and publicly funded clinical studies. Not yet recruiting: The study has not started recruiting participants; Recruiting: The study is currently recruiting participants; Active, not recruiting: The study is ongoing, and participants are receiving an intervention or being examined, but potential participants are not currently being recruited or enrolled; Terminated: The study has stopped early and will not start again; participants are no longer being examined or treated; Completed: The study has ended normally, and participants are no longer being examined or treated (that is, the last participant's last visit has occurred); Withdrawn: The study stopped early, before enrolling its first participant; Unknown: A study on ClinicalTrials.gov whose last known status was recruiting; not yet recruiting; or active, not recruiting but that has passed its completion date, and the status has not been last verified within the past 2 years).
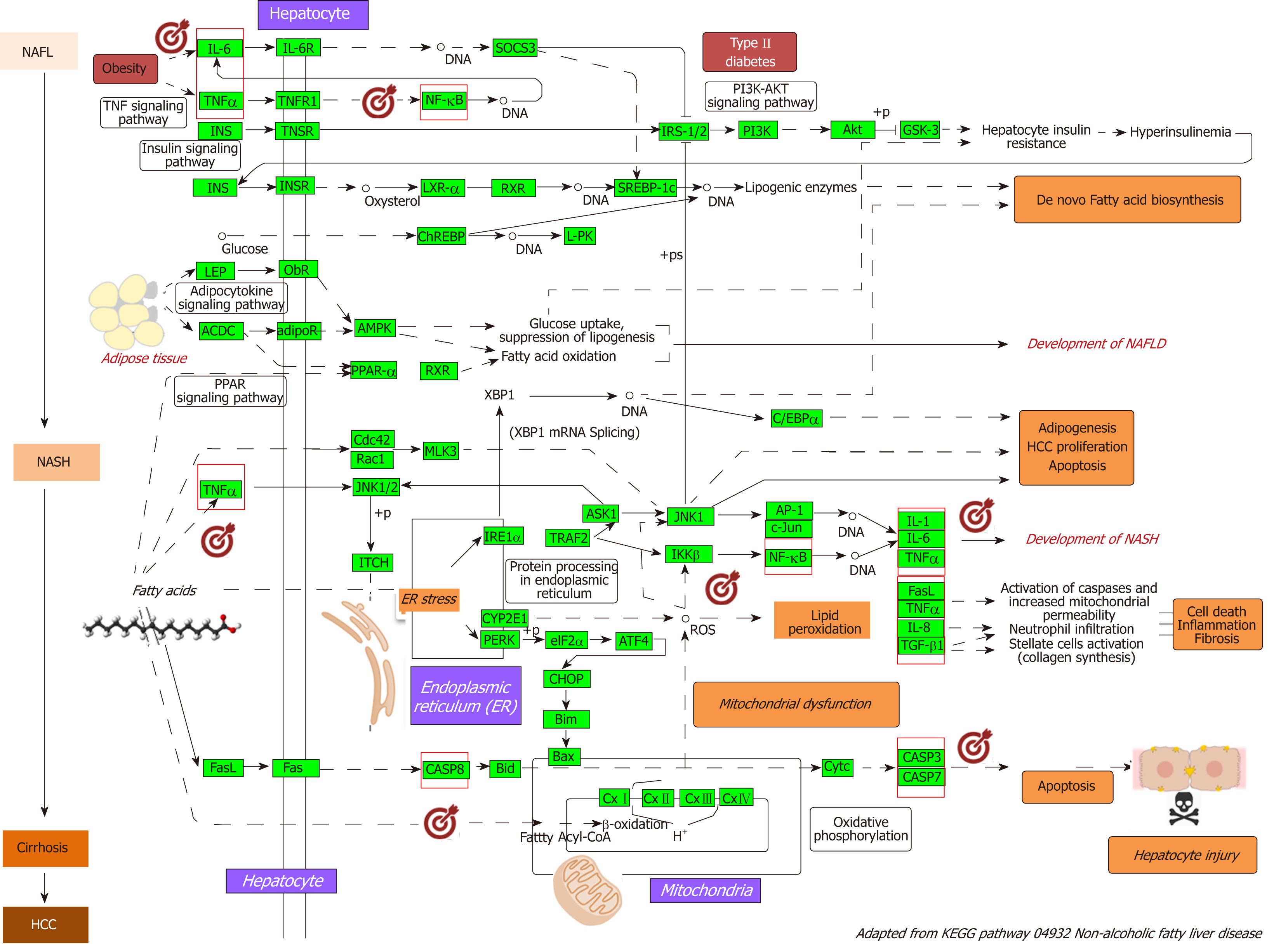
Figure 2 Nonalcoholic fatty liver disease-Kyoto Encyclopedia of Genes and Genomes pathway and mechanisms of disease pathogenesis.
Pathway was retrieved from https://http://www.genome.jp/dbget-bin/www_bget?pathway+hsa04932; figure was modified to highlight key molecular processes. This map shows a stage-dependent progression of nonalcoholic fatty liver disease (NAFLD). In the first stage of NAFLD, pathway highlights excess lipid accumulation associated with the induction of insulin resistance, which leads to a defect in insulin suppression of free fatty acids (FAAs) disposal. In addition, two transcription factors, SREBP-1c and PPARα, activate key enzymes of lipogenesis and increase the synthesis of FAAs in liver. In the second stage, pathway is presented as a consequence of the progression to nonalcoholic steatohepatitis (NASH); the production of reactive oxygen species is enhanced due to oxidation stress through mitochondrial beta-oxidation of fatty acids and endoplasmic reticulum (ER) stress, leading to lipid peroxidation. The lipid peroxidation can further cause the production of cytokines [Fas ligand, tumor necrosis factor α (TNF-α), IL-8 and transforming growth factor], promoting cell death, inflammation and fibrosis. The activation of JNK, which is induced by ER stress, TNF-α and FAAs, is also associated with NAFLD progression. Increased JNK promotes cytokine production and initiation of hepatocellular carcinoma. Major organelles involved in the pathogenesis of NASH are also highlighted in the NAFLD-pathway, including mitochondria and mitochondrial dysfunction. In the figure, molecular targets that were further selected to explore protein-chemical interactions are highlighted by red squares. NAFLD: Nonalcoholic fatty liver disease; NASH: Nonalcoholic steatohepatitis; ER: Endoplasmic reticulum; HCC: Hepatocellular carcinoma; NAFL: Nonalcoholic fatty liver; FAAs: Free fatty acids; TNFα: tumor necrosis factor α.
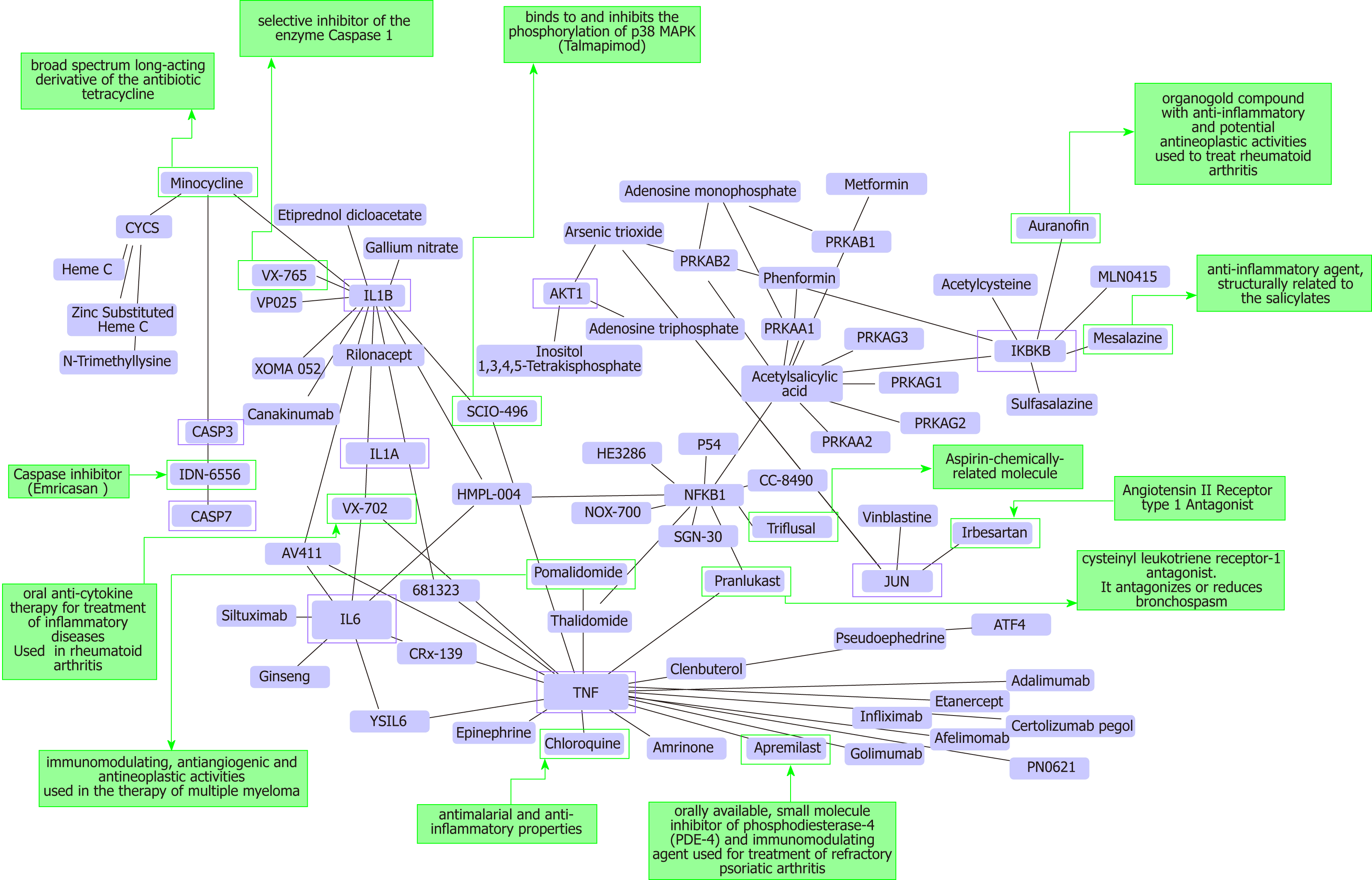
Figure 3 Protein-chemical interactions and potential repurposing drugs to target nonalcoholic steatohepatitis.
We generated a protein-chemical interaction network by mapping the significant genes/proteins that are represented in the nonalcoholic fatty liver disease-Kyoto Encyclopedia of Genes and Genomes pathway to chemicals/drugs that are annotated in the Comparative Toxicogenomics Database. The 149 genes (seeds) from our analysis were mapped to the corresponding molecular interaction database; full list of seed genes is listed in Table 1. This analysis generated a huge network composed of approximately 2000 nodes. Current figure shows chemical-drug-interactions specifically focused on selected genes/proteins of potential interest, including members of the caspase family (CASP3 and CASP7), interleukins (IL1B, IL1A, and IL6), tumor necrosis factor α (TNF-α), NFKB1 (Nuclear factor kappa B subunit 1) and IKBKB (inhibitor of nuclear factor kappa B kinase subunit beta), JUN (Jun proto-oncogene, AP-1 transcription factor subunit), AKT1 (AKT serine/threonine kinase 1). In green charts we summarized information on current use and known action of selected drugs. Interaction network was predicted by the Networkanalyst resource available at https://http://www.networkanalyst.ca/faces/home.xhtml. The network is shown as a Cytoscape graph.
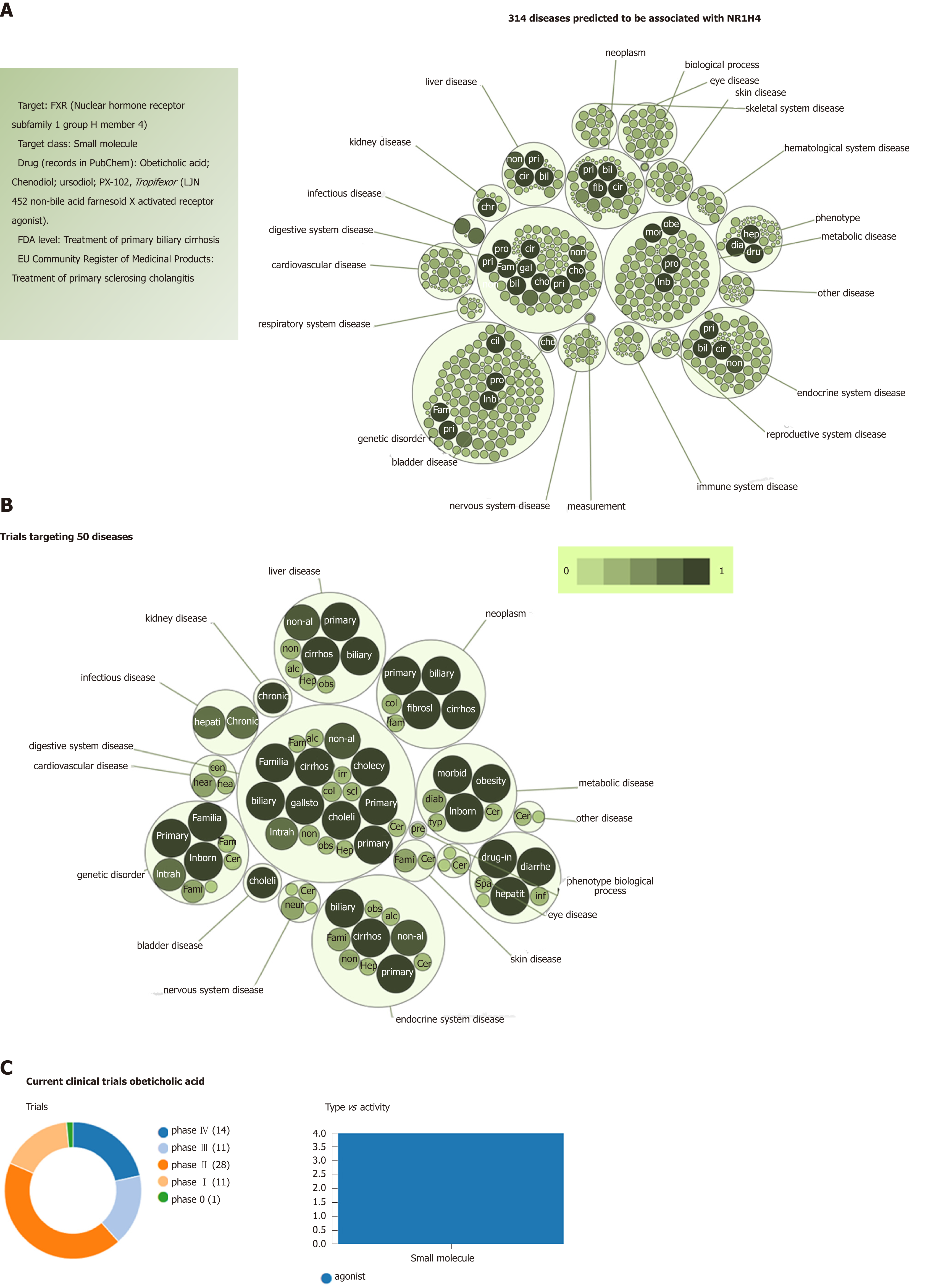
Figure 4 Farnesoid X nuclear receptor (nuclear hormone receptor subfamily 1 group H member 4): Analysis of pleiotropy.
A: Graph shows all predicted diseases associated with farnesoid X nuclear receptor; B: Clinical trials of drugs that target farnesoid X nuclear receptor. Predictions were explored in The Open Targets Platform that allows prioritisation of drug targets based on the strength of their association with a disease (https://http://http://www.targetvalidation.org/); C: Evidence curated from ClinicalTrials.gov, a database of privately and publicly funded clinical studies conducted around the world. https://clinicaltrials.gov/. Diseases are presented as bubbles grouped into therapeutic areas using their Experimental Factor Ontology relationships. The size and shade of the color of each bubble is proportional to the strength of association between the disease and farnesoid X nuclear receptor. The concept of a target-disease association is based on the analysis of several resources, including genetic associations (GWAS Catalog, UniProt, European Variation Archive, Gene2Phenotype), somatic mutations (Cancer Gene Census, European Variation Archive somatic, IntOGen), RNA expression (expression atlas), drugs (ChEMBL), affected pathways (Reactome), animal models (PhenoDigm) and text mining (Europe PMC). The platform is available at https://http://www.targetvalidation.org. Data last updated December 2018.
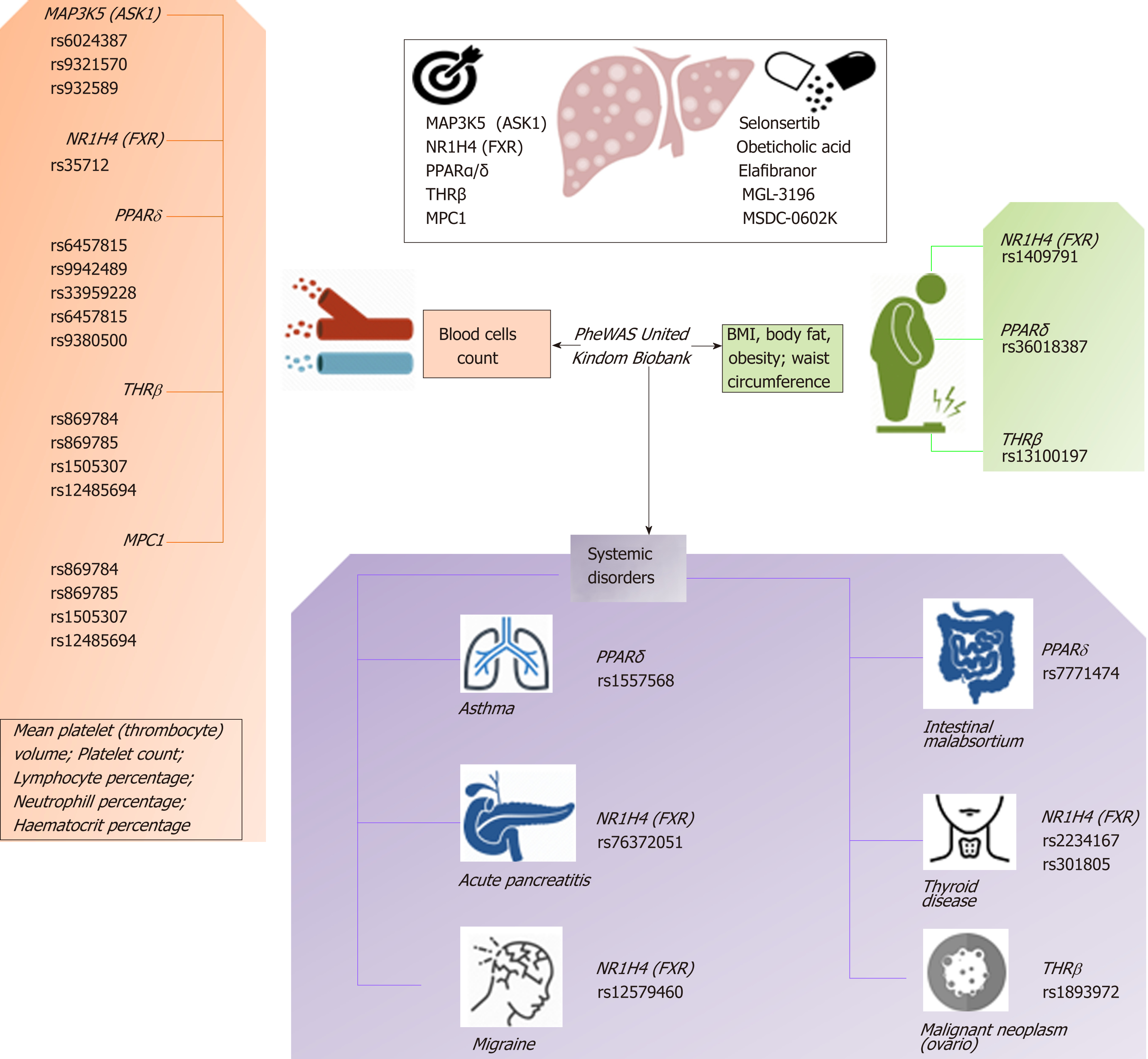
Figure 5 The complexity of molecular targets and novel nonalcoholic steatohepatitis drugs: Pleiotropy assessed in the PheWAS United Kindom Biobank.
Figure shows associations between gene variants in five nonalcoholic steatohepatitis-related molecular targets (MAP3K5/ASK1, FXR, PPARα/δ, THRβ, and MPC1) with different traits and phenotypes in the UK-PheWAS (Phenome-wide association study). Information regarding single nucleotide polymorphisms and associations were retrieved from the United Kindom Biobank (http://geneatlas.roslin.ed.ac.uk/).
- Citation: Sookoian S, Pirola CJ. Repurposing drugs to target nonalcoholic steatohepatitis. World J Gastroenterol 2019; 25(15): 1783-1796
- URL: https://www.wjgnet.com/1007-9327/full/v25/i15/1783.htm
- DOI: https://dx.doi.org/10.3748/wjg.v25.i15.1783