Copyright
©The Author(s) 2018.
World J Gastroenterol. Aug 14, 2018; 24(30): 3374-3383
Published online Aug 14, 2018. doi: 10.3748/wjg.v24.i30.3374
Published online Aug 14, 2018. doi: 10.3748/wjg.v24.i30.3374
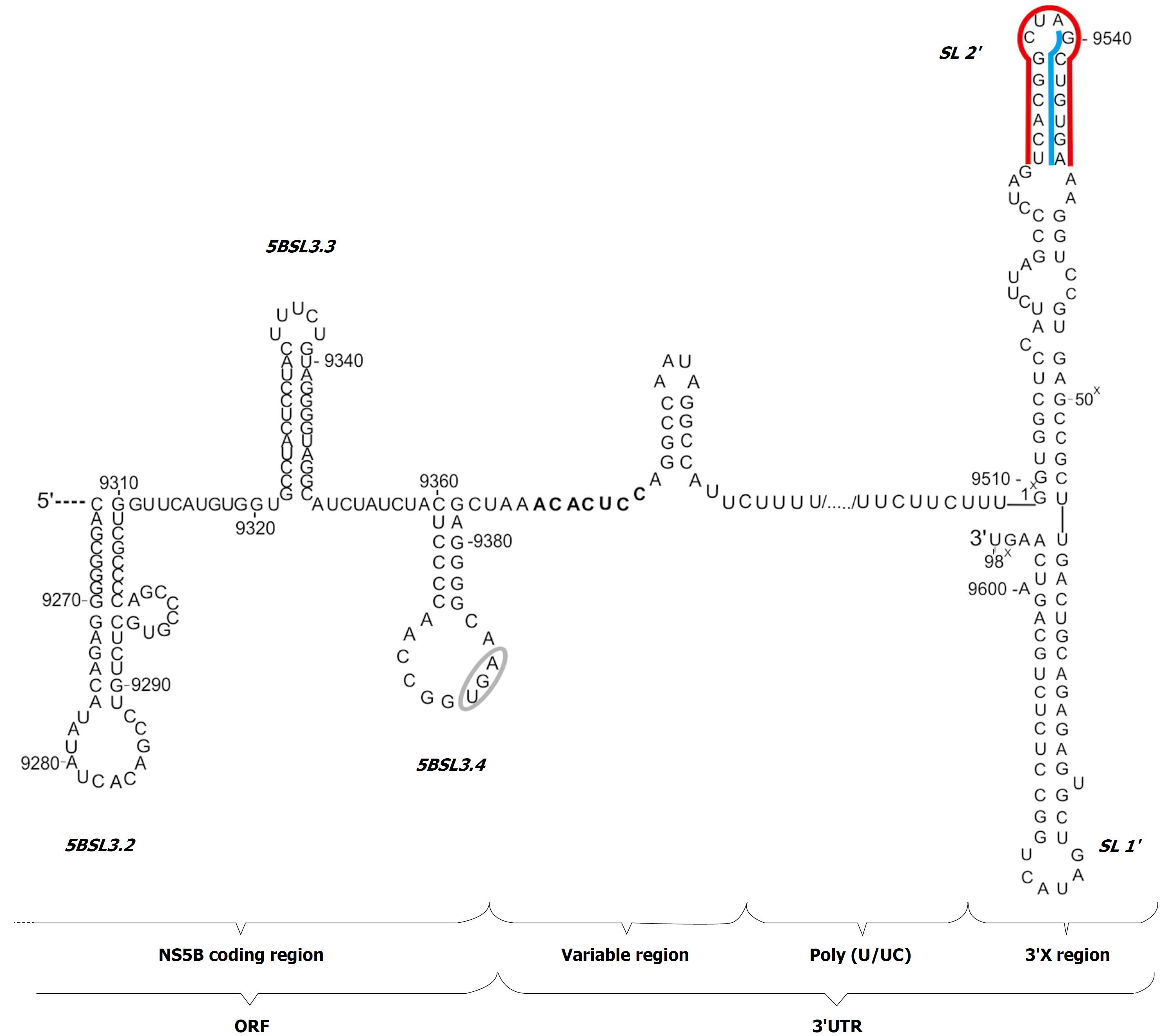
Figure 1 Secondary structure model of the 3'UTR of hepatitis C virus genome with adjacent 3' terminal sequence of the coding region.
The stop codon is indicated by a gray ellipse. The regions involved in the kissing-interactions are marked with blue lines; the region involved in dimerization is indicated by a red line; the seed region for miR122 is highlighted in bold.
Figure 2 Diverse secondary structure models proposed for the 3'X region of hepatitis C virus genome.
A: The 3xSL model[17,18]; B: The 4xSL model[20]; C: The 2xSL model[21]. The region involved in the kissing-interactions is indicated with blue line, the region involved in dimerization is marked with red line; possible alternative folding of separate fragments are displayed as gray rectangles.
Figure 3 Long-range RNA-RNA interactions proposed for the 3'X region of hepatitis C virus genome.
A: The homodimeric interactions between two 3'X regions embedded into two RNA molecules, model according to Cantero-Camacho et al[24]; B: The kissing-interactions with SL5B3.2[22,28,31,33]. Nucleotide sequence involved in dimerization is additionally indicated with red line, while those involved in the kissing-interactions are marked with blue lines.
- Citation: Dutkiewicz M, Ciesiołka J. Form confers function: Case of the 3’X region of the hepatitis C virus genome. World J Gastroenterol 2018; 24(30): 3374-3383
- URL: https://www.wjgnet.com/1007-9327/full/v24/i30/3374.htm
- DOI: https://dx.doi.org/10.3748/wjg.v24.i30.3374