Copyright
©The Author(s) 2018.
World J Gastroenterol. Apr 14, 2018; 24(14): 1507-1520
Published online Apr 14, 2018. doi: 10.3748/wjg.v24.i14.1507
Published online Apr 14, 2018. doi: 10.3748/wjg.v24.i14.1507
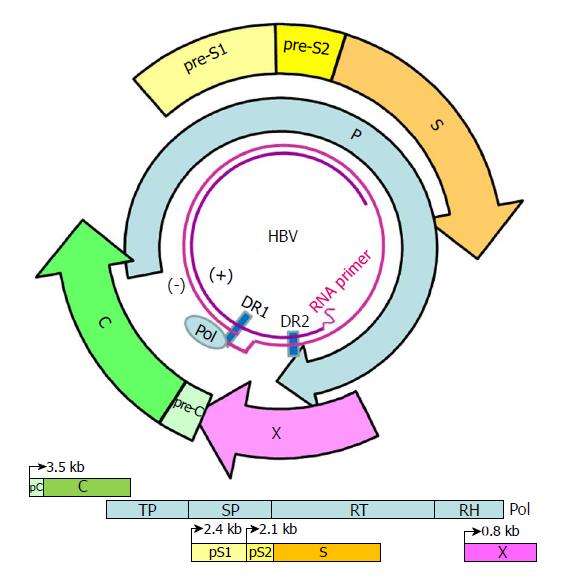
Figure 1 Genome structure and organization of hepatitis B virus.
The relaxed-circular DNA genome of HBV with a complete minus strand and incomplete plus strand is shown (inner circle), along with the four main open reading frames (ORFs): pre-S/S; precore/core (pC/C); Pol, including four domains: TP, SP, reverse transcriptase (RT), and RNase H (RH); and X. The minus (-) and plus (+) DNA strands are marked. The HBV Pol and capped mRNA oligomer at the 5' end of the (-) and (+) strands as well as the DR-1 and DR-2 are illustrated. The space between the DR-1 and DR-2 is the "cohesive overlap region." The (+) strand is typically incomplete.
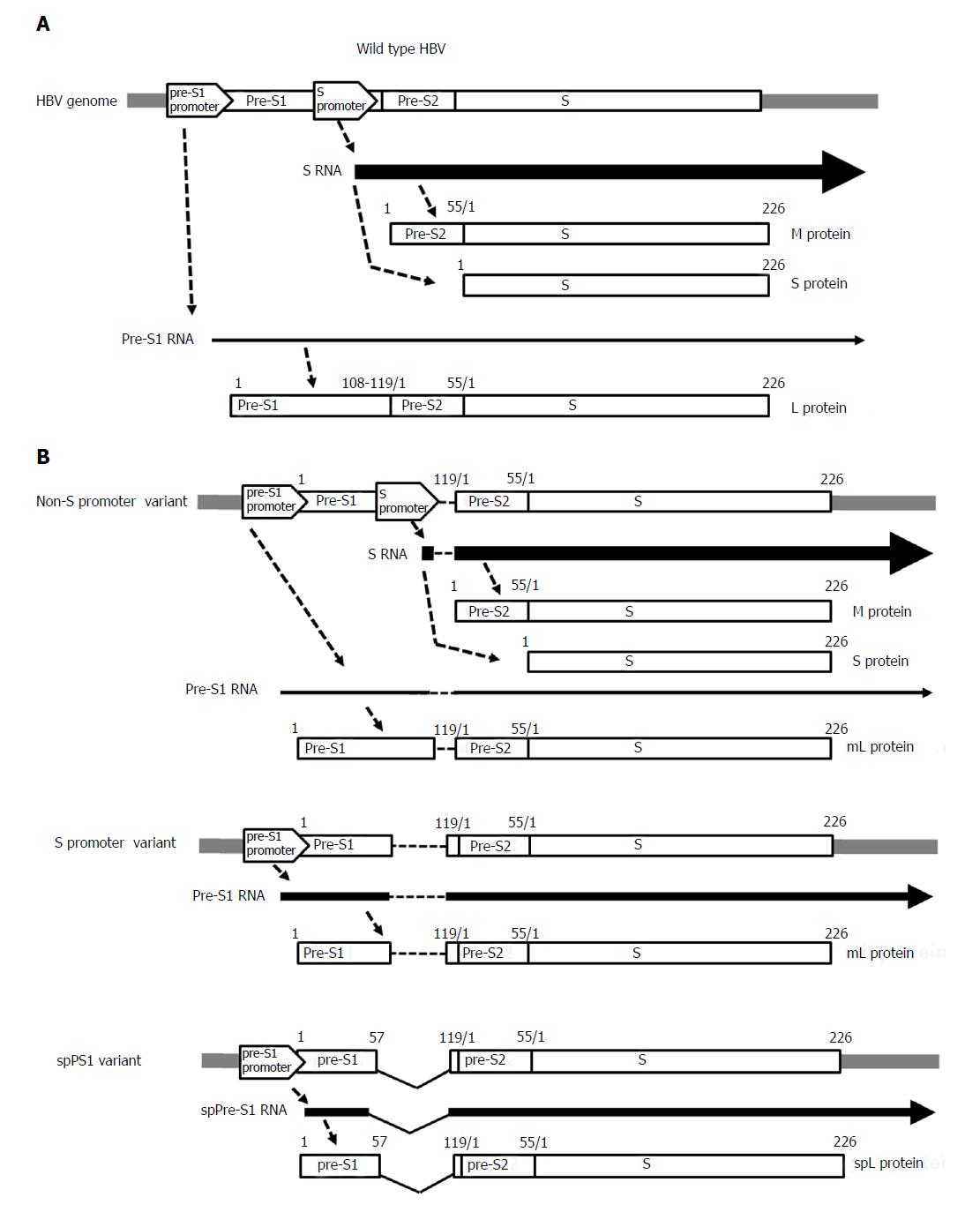
Figure 2 Gene expression of the pre-S/S gene in (A) wild-type hepatitis B virus and (B) pre-S/S variants: non-S promoter, S promoter, and spPS1.
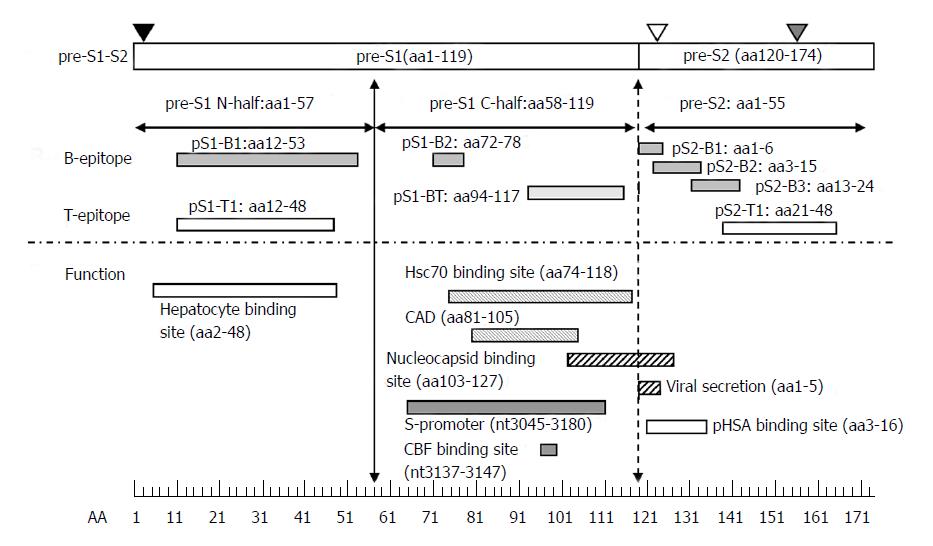
Figure 3 Immune epitopes and functional domains within the hepatitis B virus pre-S region.
The pre-S region consists of the pre-S1 and pre-S2 regions. The pre-S1 region contains 119 amino acids in HBV genotypes B or C and is further divided into two parts: the N half (amino acids 1-57) and C half (amino acids 58-119). The pre-S2 region contains 55 amino acids. The pre-S domain contains many B- or T-epitopes and exerts multiple functions, as illustrated. The N-half of pre-S1 contains a hepatocyte binding site essential for infection. The C-half of pre-S1 contains a heat-shock protein 70 (Hsc70) binding site and cytosolic anchorage determinant (CAD) vital for dual topology of L proteins as well as a nucleocapsid binding site (NBS) for virion morphogenesis. The C-half of pre-S1 also contains an S-promoter and CCAAT binding factor (CBF) binding site necessary for expression of the S gene. The pre-S2 region has a polymerized human serum albumin (pHSA) binging site and viral secretion (VS) site. Black triangle, myristylation at second amino acid; white triangle, N-link glycosylation at N-4 of the M protein; gray triangle, O-link glycosylation at T-37 of the M protein. B-epitopes: pS1-B1, pS1-B2, pS2-B1, pS2-B2, and pS2-B3. T-epitopes: pS1-T1 and pS2-T1. B- and T-epitope: pS1-BT.
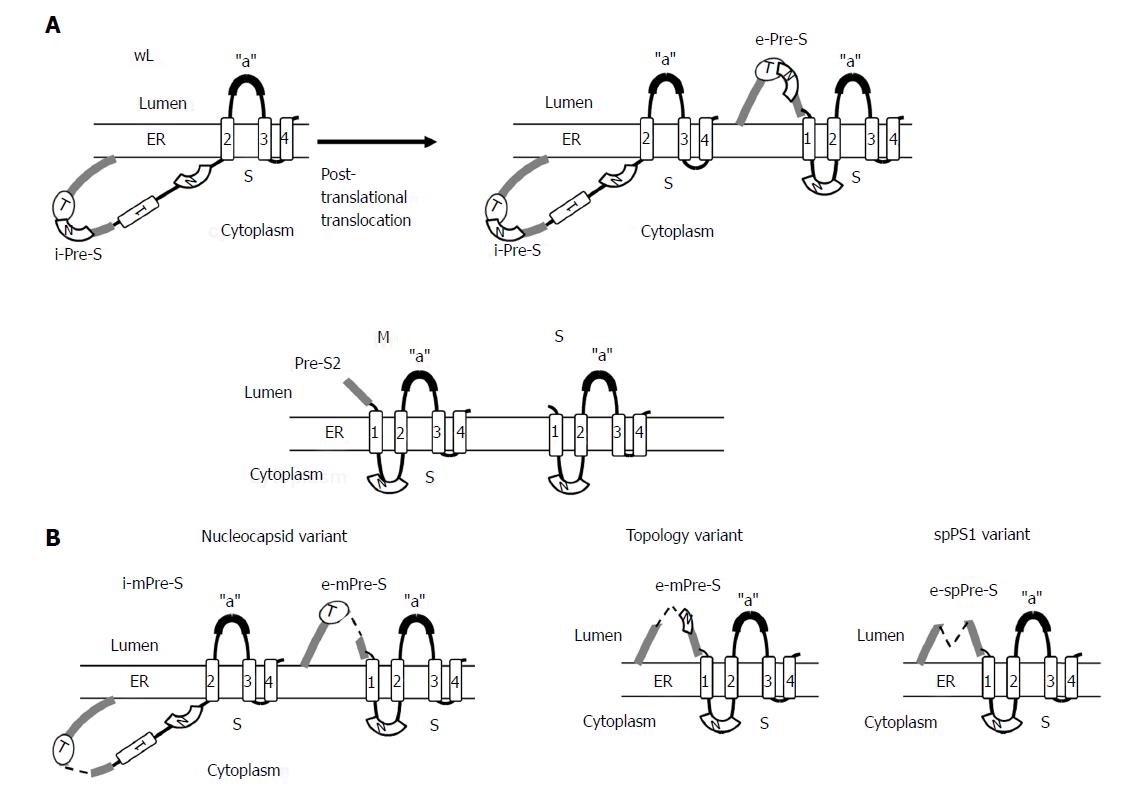
Figure 4 Topology of wild-type small (S), medium (M), and large (L) envelope proteins.
The predicted four membrane-spanning segments (indicated by rectangular boxes) of S project their N and C termini into the endoplasmic reticulum lumen (A). The M proteins exhibit a topology similar to the S protein with their N-terminal pre-S2 domain protruding into the endoplasmic reticulum (ER) lumen, whereas the L proteins display a dual topology. Upon cotranslational membrane integration, the pre-S domains of L proteins are initially located on the cytosolic side of the ER membrane (i-Pre-S); they are controlled by the dual topology site (indicated by an oval). During maturation (marked by the arrow), nearly half of mature L-protein molecules posttranslationally translocate their pre-S region to the luminal space (e-Pre-S). The nucleocapsid (N) binding sites in the pre-S and S region are indicated by the white curved box. (B) The L-protein topology of pre-S/S variants. The nucleocapsid variant demonstrates a dual topology, and the topology variants and spPS1 variants display a uniform topology. The broken line indicates deletion, and “a” indicates “a” determinant.
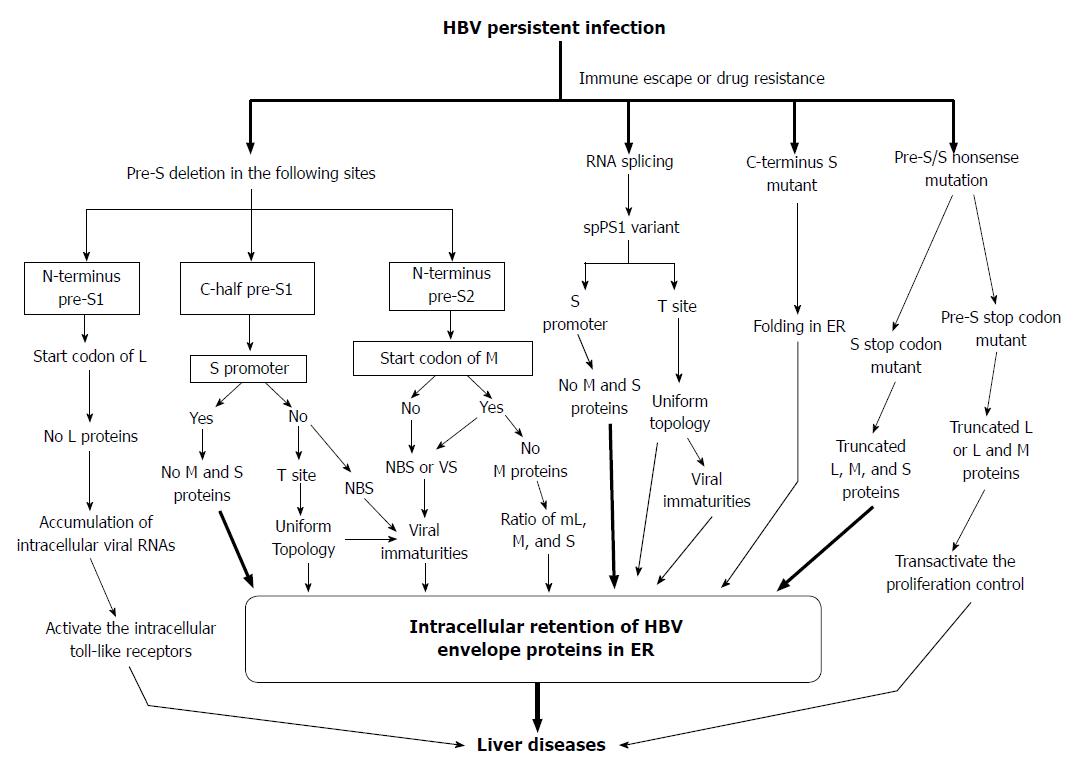
Figure 5 Proposed model for the generation of pre-S/S variants and their possible roles in liver damage and carcinogenesis.
HBV: Hepatitis B virus; ER: Endoplasmic reticulum; NBS: Nucleocapsid binding site; VS: Viral secretion.
- Citation: Chen BF. Hepatitis B virus pre-S/S variants in liver diseases. World J Gastroenterol 2018; 24(14): 1507-1520
- URL: https://www.wjgnet.com/1007-9327/full/v24/i14/1507.htm
- DOI: https://dx.doi.org/10.3748/wjg.v24.i14.1507