Copyright
©The Author(s) 2016.
World J Gastroenterol. Jan 28, 2016; 22(4): 1487-1496
Published online Jan 28, 2016. doi: 10.3748/wjg.v22.i4.1487
Published online Jan 28, 2016. doi: 10.3748/wjg.v22.i4.1487
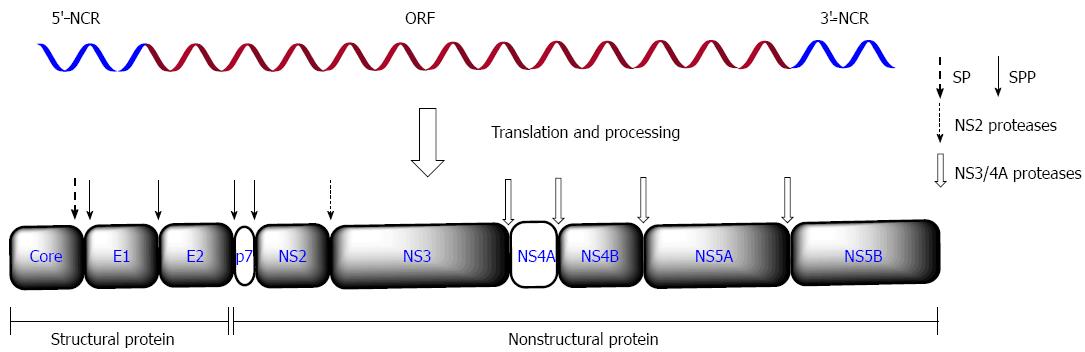
Figure 1 Hepatitis C virus genome structure and processing of polyprotein.
Hepatitis C virus (HCV) genome is a positive-sense, single-stranded RNA about 9.6 kb with a conserved 5’-NCR, one ORF and a conserved 3’-NCR. The polyprotein is cleaved by host ER signal peptidase (SP) or signal peptide peptidase (SPP) into three structural proteins (core, E1 and E2) and by the viral NS2 and/or NS3/4A protease into seven nonstructural proteins (p7, NS2, NS3, NS4A, NS4B, NS5A and NS5B). Arrows denote the cleavage sites of host or virus proteases. NCR: Noncoding region; ORF: Open reading frame; ER: Endoplasmic reticulum.
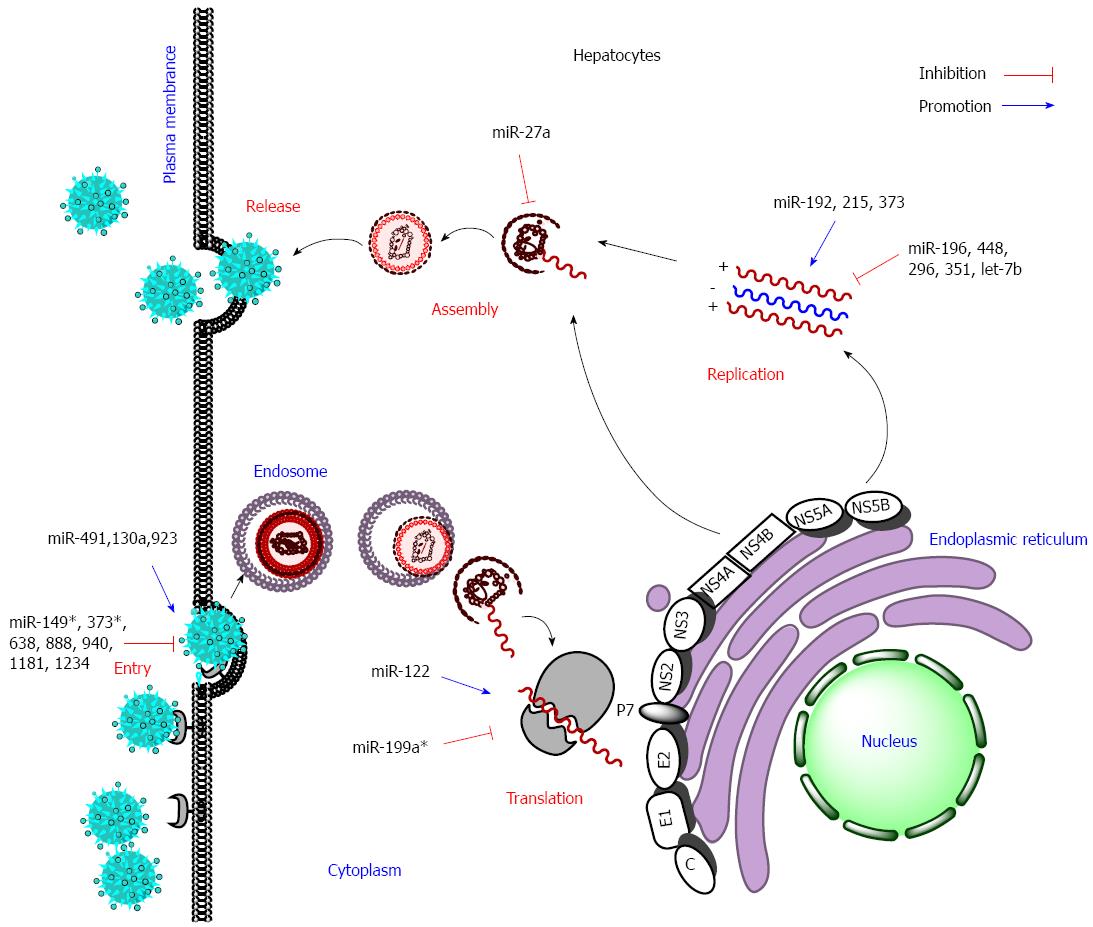
Figure 2 Effect of some cellular miRNAs on hepatitis C virus life cycle.
Host miRNAs inhibit or promote hepatitis C virus (HCV) replication through targeting entry, translation, replication, or assembly. Red blunt arrows denote anti-HCV effects and the blue solid arrows denote promotion effects on HCV replication.
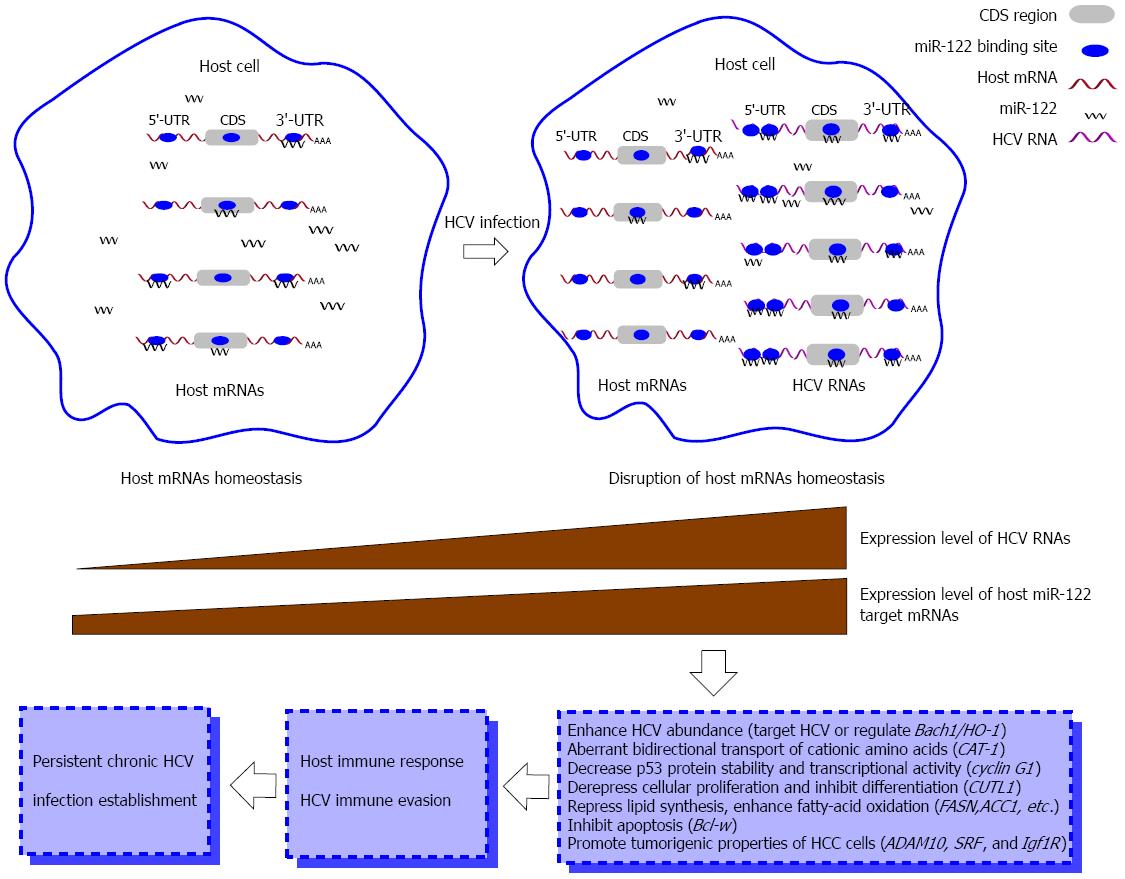
Figure 3 miR-122-centered cvhRNA network disrupts host homeostasis and establishes persistent hepatitis C virus infection.
Hepatitis C virus (HCV) RNAs harbor miR-122 binding sites in the 5’ and 3’-UTRs and coding sequence regions and thus efficiently sequester cellular miR-122 from binding to its host target mRNAs. In this manner, HCV RNAs de-repress expression of host target mRNAs, such as CAT-1, cyclin G1 and CUTL1, alter host homeostasis, and partly cause a host antiviral response. The interplay between host and HCV finally forms persistent chronic HCV infection. UTR: Untranslated region; CDS: Coding sequence.
- Citation: Li H, Jiang JD, Peng ZG. MicroRNA-mediated interactions between host and hepatitis C virus. World J Gastroenterol 2016; 22(4): 1487-1496
- URL: https://www.wjgnet.com/1007-9327/full/v22/i4/1487.htm
- DOI: https://dx.doi.org/10.3748/wjg.v22.i4.1487