Copyright
©The Author(s) 2016.
World J Gastroenterol. Oct 7, 2016; 22(37): 8283-8293
Published online Oct 7, 2016. doi: 10.3748/wjg.v22.i37.8283
Published online Oct 7, 2016. doi: 10.3748/wjg.v22.i37.8283
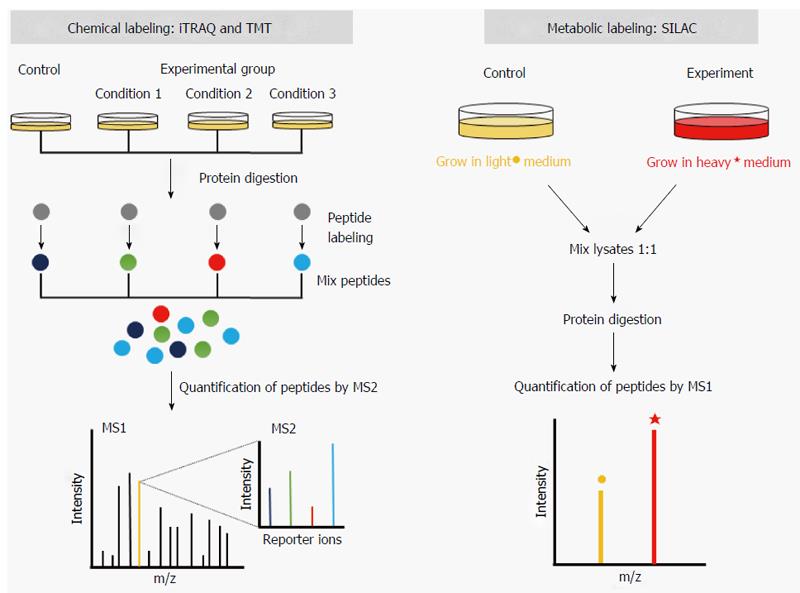
Figure 1 Schematic overview of labeling strategies used in quantitative proteomics.
Chemical labeling utilizes the isobaric tags for relative and absolute quantification (iTRAQ) and the tandem mass tags (TMT). In this approach, proteolytic peptides from separate samples are labeled with discrete isobaric tags and pooled. Precursor peptide ions are fragmented (MS2) to generate reporter ions with distinct m/z, whose relative intensities represent the relative abundances of the peptides producing the corresponding reporter ion. Metabolic labeling represented by the stable isotope labeling with amino acids in cell culture (SILAC) strategy takes advantage of the metabolic incorporation of heavy amino acids into mature proteins. In this strategy, the relative peak intensities (MS1) represent the abundances of the precursor peptide ions.
- Citation: Kang C, Lee Y, Lee JE. Recent advances in mass spectrometry-based proteomics of gastric cancer. World J Gastroenterol 2016; 22(37): 8283-8293
- URL: https://www.wjgnet.com/1007-9327/full/v22/i37/8283.htm
- DOI: https://dx.doi.org/10.3748/wjg.v22.i37.8283