Copyright
©The Author(s) 2016.
World J Gastroenterol. Aug 21, 2016; 22(31): 7030-7045
Published online Aug 21, 2016. doi: 10.3748/wjg.v22.i31.7030
Published online Aug 21, 2016. doi: 10.3748/wjg.v22.i31.7030
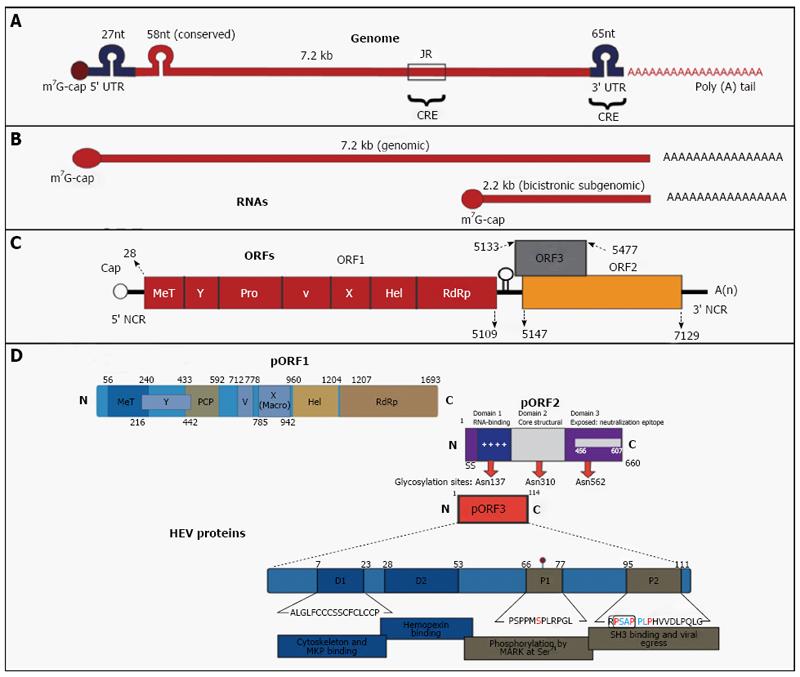
Figure 1 Hepatitis E virus.
Genomic organization. A: The hepatitis E virus genome; B: Genomic RNA and bicistronic subgenomic RNA; C: Open reading frames (ORFs) and (D) 3 encoded proteins (pORF1, pORF2 and pORF3). For details, see text about hepatitis E virus. Adopted from Khuroo et al[143], 2016.
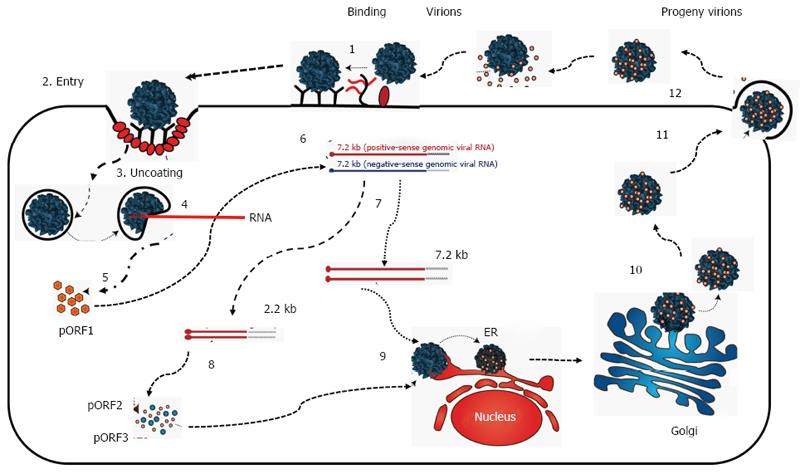
Figure 2 Proposed replication of hepatitis E virus.
For details, see text about hepatitis E replication. Adopted from Khuroo et al[143], 2016.
Figure 3 Evolutionary history of hepatitis E virus.
The times to the most recent common ancestors (tMRCAs) for all four genotypes of HEV were calculated using BEAST to conduct a Bayesian analysis of HEV. The population dynamics for genotypes 1, 3 and 4 were analyzed using skyline plots. For details see text on HEV evolution. Source of data Purdy et al[50] 2010. HEV: Hepatitis E virus.
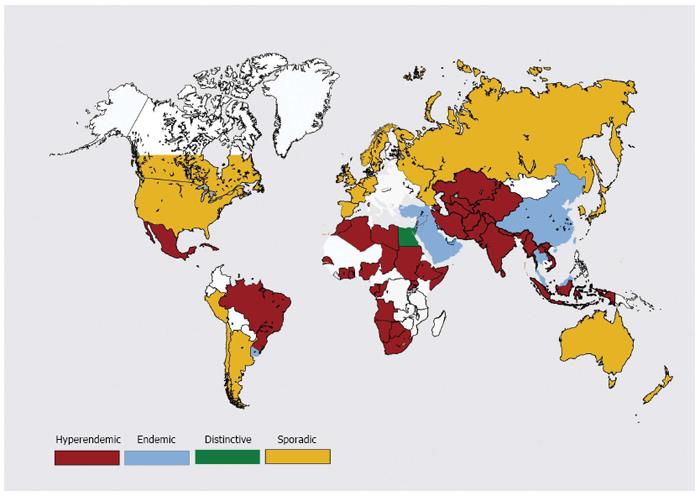
Figure 4 Global distribution of hepatitis E disease.
See text under “global distribution” for explanation.
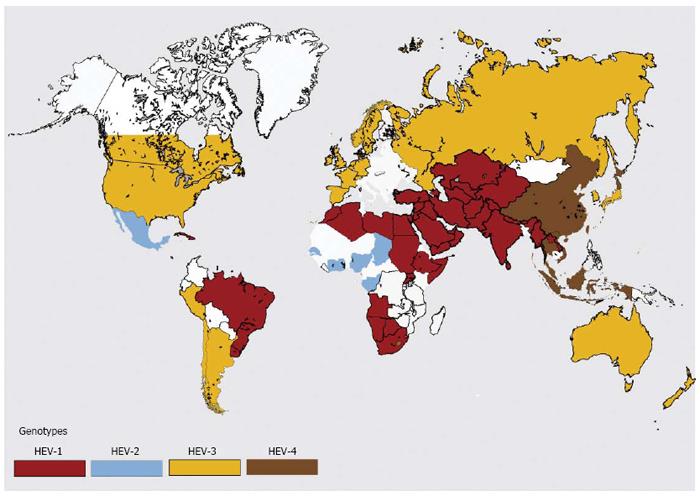
Figure 5 Global distribution of hepatitis E virus genotypes.
See text under “global distribution” for explanation.
- Citation: Khuroo MS, Khuroo MS, Khuroo NS. Hepatitis E: Discovery, global impact, control and cure. World J Gastroenterol 2016; 22(31): 7030-7045
- URL: https://www.wjgnet.com/1007-9327/full/v22/i31/7030.htm
- DOI: https://dx.doi.org/10.3748/wjg.v22.i31.7030