Copyright
©The Author(s) 2015.
World J Gastroenterol. Nov 7, 2015; 21(41): 11597-11608
Published online Nov 7, 2015. doi: 10.3748/wjg.v21.i41.11597
Published online Nov 7, 2015. doi: 10.3748/wjg.v21.i41.11597
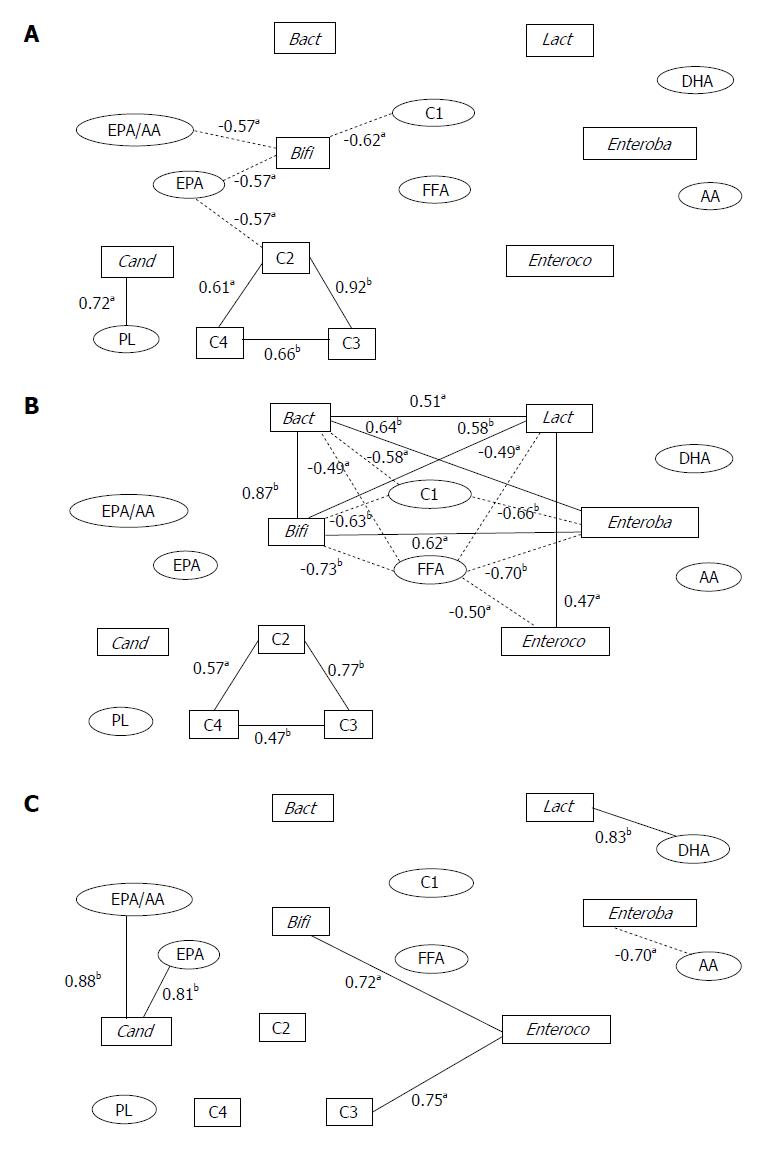
Figure 1 Correlation networks among fecal microflora and organic acid and serum organic and fatty acid concentrations in hepatic cancer patients.
Square boxes indicate fecal components and ellipsoids indicate serum components. Solid lines indicate positive correlations and dotted lines indicate negative correlations. A: Normal liver group; B: Chronic hepatitis or liver fibrosis group; C: Liver cirrhosis group. aP < 0.05 and bP < 0.01 by Pearson’s correlation coefficient test. Bact: Bacteroidaceae; Bifi: Bifidobacterium; Lact: Lactobacillus; Enteroba: Enterobacteriaceae; Enteroco: Enterococcus; Cand: Candida; C1: Formic acid; C2: Acetic acid; C3: Propionic acid; C4: Butyric acid; AA: Arachidonic acid; EPA: Eicosapentaenoic acid; DHA: Docosahexaenoic acid; FFA: Free fatty acid; PL: Phospholipid. Data adapted from Usami et al[46].
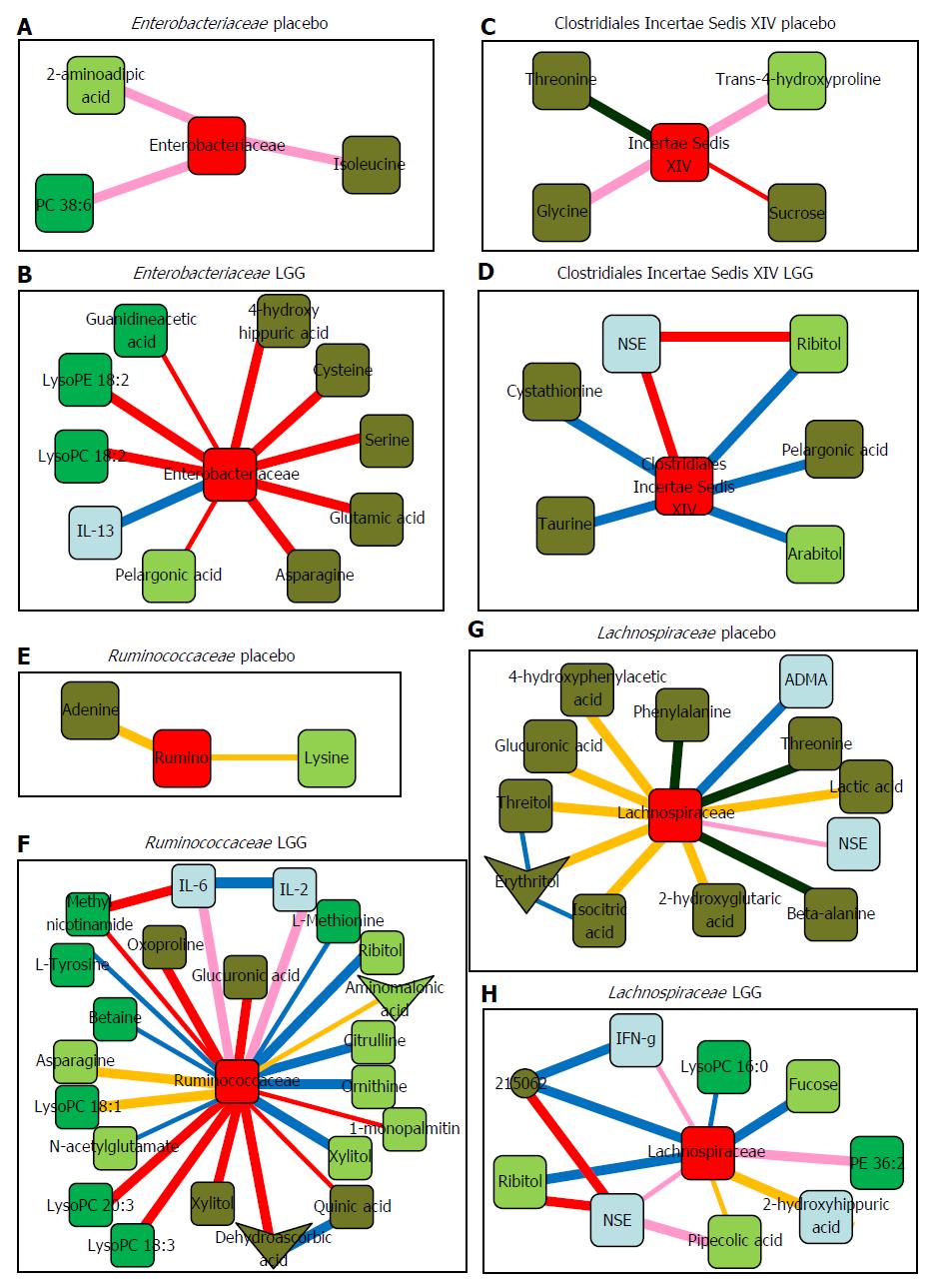
Figure 2 Sub-networks showing correlation network differences from baseline to week 8 in placebo and in Lactobacillus GG groups separately centered on selected bacterial taxa.
Color of nodes: Blue, inflammatory cytokine; light green, serum metabolites; dark green, urine metabolites. Color of edges: pink, negative remained negative but there is a net loss of negative correlation; dark blue, negative changed to positive; yellow, positive remained positive but there is a net loss of positive correlation; red, positive to negative; dark green, complete shift of negative to positive; military green, complete shift of positive to negative. A, B: Sub-networks of correlation changes centered around Enterobacteriaceae; C, D: Sub-networks of correlation changes centered around Clostridiales Incertae Sedis XIV; E, F: Sub-networks of correlation changes centered around Ruminococcaceae; G, H: Sub-networks of correlation changes centered around Lachnospiraceae. NSE: Neuron-specific enolase; IL: Interleukin; IFN: Interferon; LysoPC: Lysophosphatidylcholine; LysoPE: Lysophosphatidylethanolamine; ADMA: Asymmetricdimethylarginine; Rumino: Ruminococcaceae. Data adapted from Bajaj et al[69].
- Citation: Usami M, Miyoshi M, Yamashita H. Gut microbiota and host metabolism in liver cirrhosis. World J Gastroenterol 2015; 21(41): 11597-11608
- URL: https://www.wjgnet.com/1007-9327/full/v21/i41/11597.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i41.11597