Copyright
©The Author(s) 2015.
World J Gastroenterol. Jan 21, 2015; 21(3): 868-877
Published online Jan 21, 2015. doi: 10.3748/wjg.v21.i3.868
Published online Jan 21, 2015. doi: 10.3748/wjg.v21.i3.868
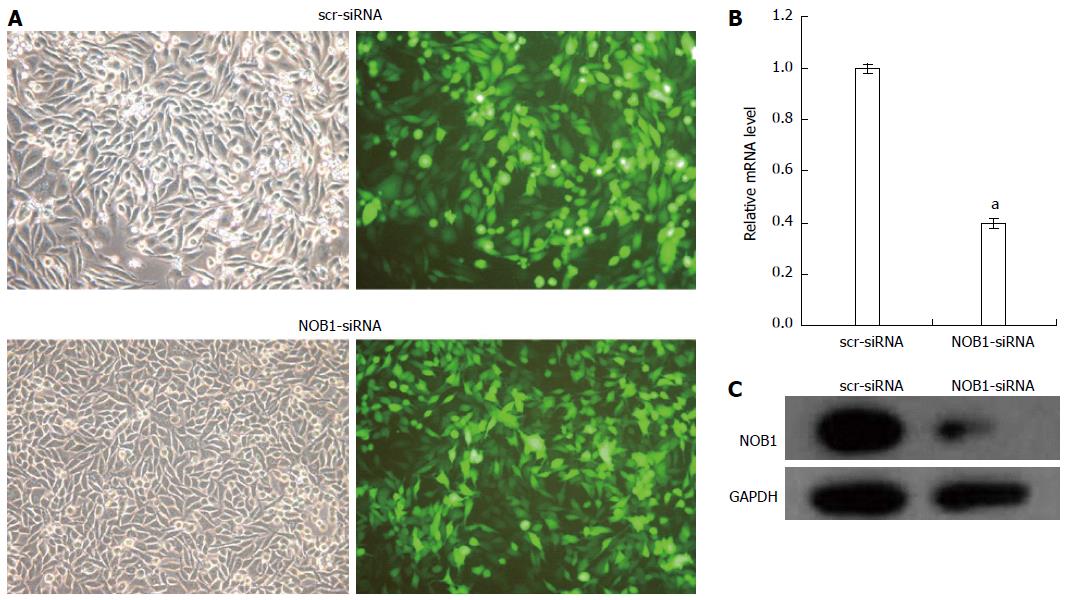
Figure 1 Lentivirus-mediated siRNA decreased NOB1 expression in RKO cells.
A: Infection efficiency was estimated 4 d after infection at MOI of 10. Green fluorescent protein (GFP) expression in infected cells was observed under light and fluorescence microscopy, respectively. Light micrograph (left); Fluorescent micrograph (right) (× 200); B: Total RNA was extracted 5 d after infection, and relative NOB1 mRNA expression was determined by qRT-PCR. GAPDH expression was used as a loading control. Data are presented as mean ± SD of three independent experiments. aP < 0.05 vs scr-siRNA; C: Total cellular protein was extracted 7 d after infection and determined by western blot analysis using antibodies against NOB1. GAPDH was used as an internal control. MOI: Multiplicity of infection; GFP: Green fluorescent protein; GAPDH: Glyceraldehyde-3-phosphate dehydrogenase.
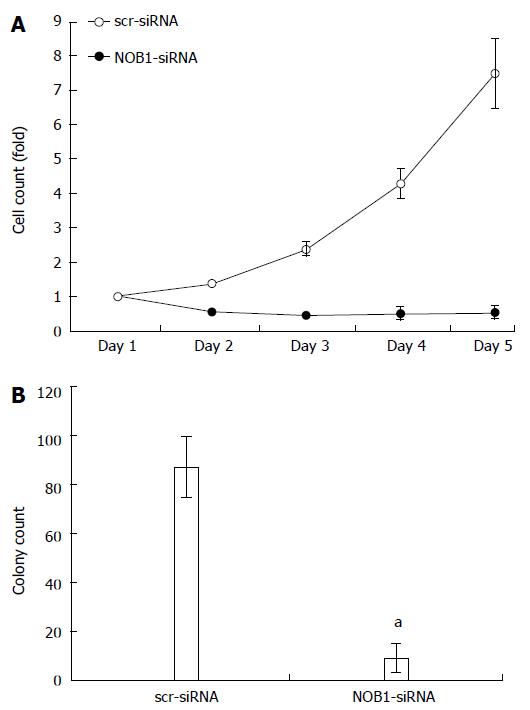
Figure 2 Effect of NOB1 knockdown on cell growth in RKO cells.
A: The infected cells expressing GFP were imaged and counted using the Cellomics ArrayScanTM High Content Screening (HCS) Reader once a day for 5 d. Cell growth curves in scr-siRNA and NOB1-siRNA infected RKO cells are shown; B: Cell growth was detected by the colony formation assay in RKO cells 14 d after infection of scr-siRNA and NOB1-siRNA cells. RKO cells were seeded at 500 cells/well and allowed to form colonies. Cell colonies were imaged and counted using the Cellomics ArrayScanTM HCS Reader. Data are presented as mean ± SD of three independent experiments. aP < 0.05 vs scr-siRNA. scr-siRNA: Cells infected with lentivirus-mediated scramble small interfering RNA; NOB1-siRNA: Cells infected with lentivirus-mediated NOB1-siRNA; GFP: Green fluorescent protein; qRT-PCR: Quantitative reverse transcription polymerase chain reaction.
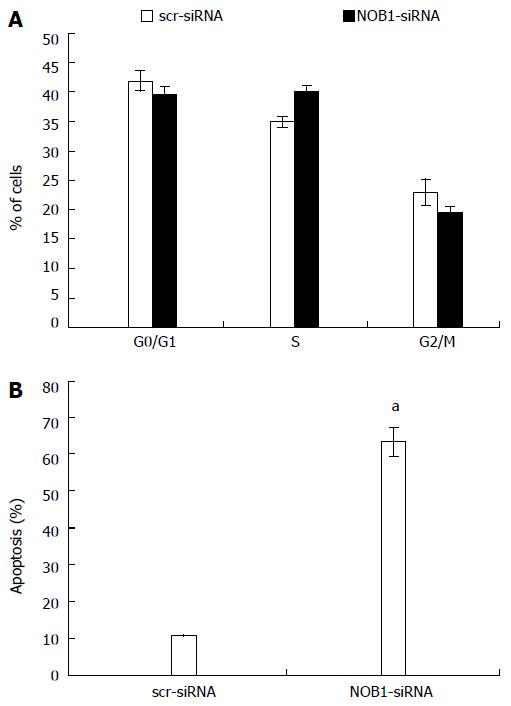
Figure 3 Effects of NOB1 downregulation on cell cycle progression and apoptosis in RKO cells.
A: Cell cycle distribution was determined by flow cytometric analysis at day 6 of infection; B: Flow cytometric analysis of cell apoptosis was performed on day 7 of infection. Data are presented as mean ± SD of three independent experiments. aP < 0.05 vs scr-siRNA. scr-siRNA: Cells infected with lentivirus-mediated scramble small interfering RNA; NOB1-siRNA: Cells infected with lentivirus-mediated NOB1-siRNA.
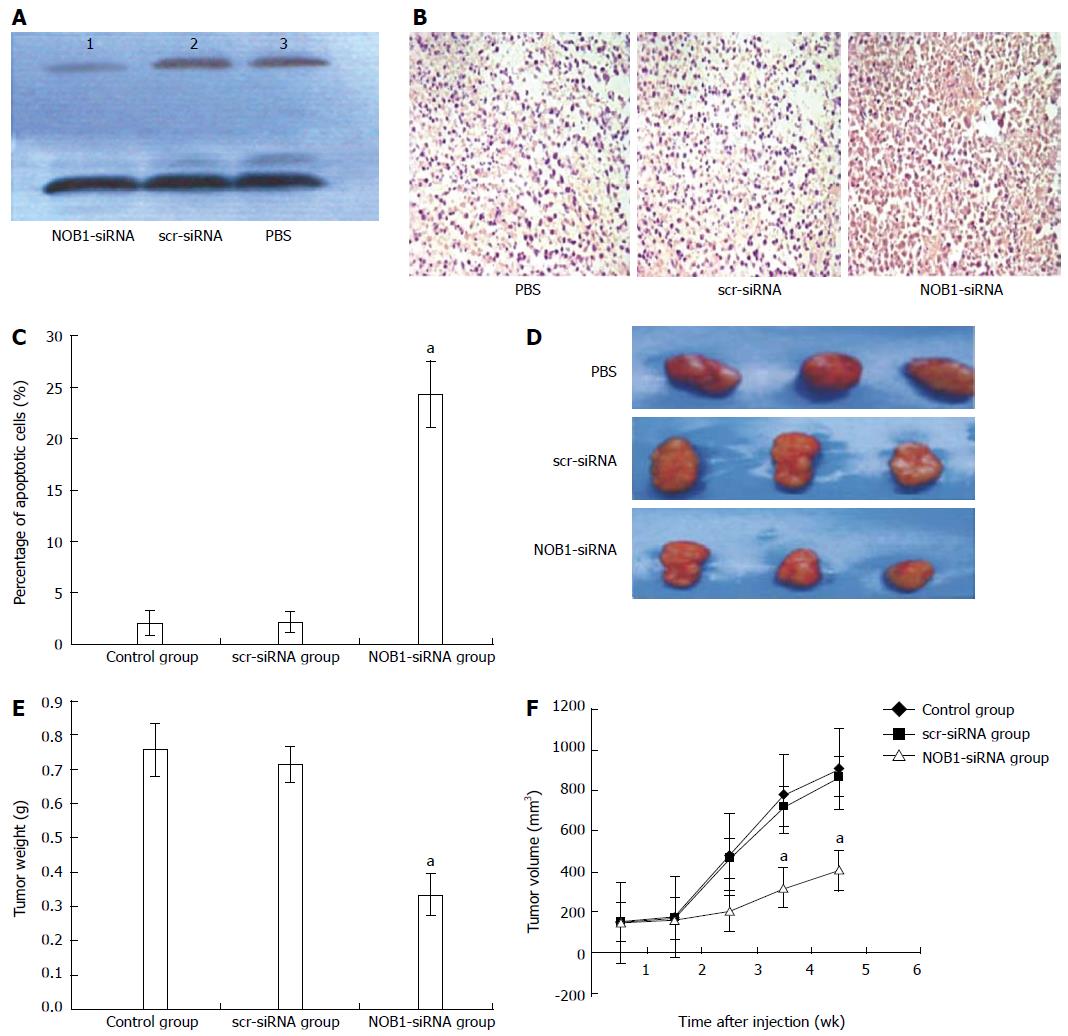
Figure 4 Suppression of tumor growth by lentivirus-mediated NOB1-siRNA in RKO cell xenograft mouse model.
A: RKO cell xenograft mice were injected intratumorally with lentivirus-mediated NOB1-siRNA, scr-siRNA, or PBS. The size of the primary tumors was measured every week. Mice were sacrificed after 5 wk. NOB1-siRNA reduced NOB-1 expression in xenografted tumor tissue as determined by Western blot; B: NOB1-siRNA induced significant apoptosis in xenografted tumor tissue detected by TUNEL staining as shown in representative images; C: Plotted percentages of apoptotic cells; D: Typical pictures from RKO cell xenografted tumors infected by PBS, src-RNA and NOB1-siRNA; E: Plotted tumor weight; F: Tumor volume in RKO cell xenograft mouse model after NOB1-siRNA infection. aP < 0.05 vs scr-siRNA. scr-siRNA: Cells infected with lentivirus-mediated scramble small interfering RNA; NOB1-siRNA: Cells infected with lentivirus-mediated NOB1-siRNA; PBS: Phosphate buffered saline.
Figure 5 Heat map showing 56 genes differentially expressed in NOB1-siRNA infected RKO cells and enriched in the cancer pathway.
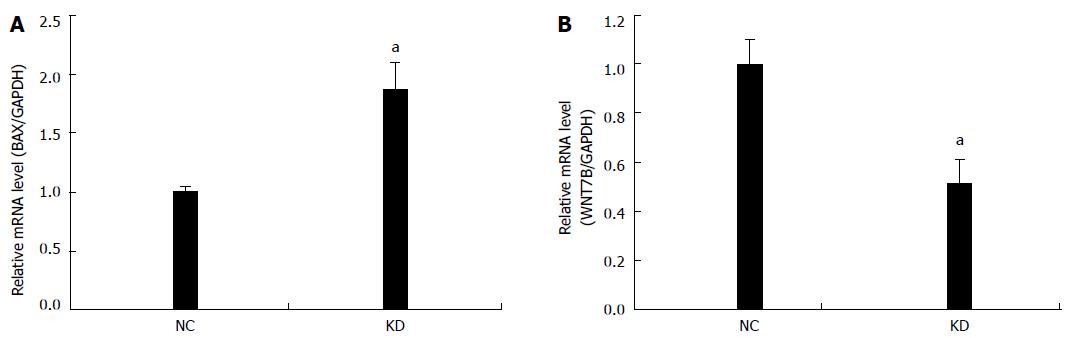
Figure 6 Quantitative reverse transcription polymerase chain reaction demonstrated NOB1-siRNA up-regulating BAX and down-regulating WNT7B mRNA in RKO cells.
GAPDH expression was used as a loading control. Data are presented by the mean ± SD of three independent experiments. aP < 0.05 vs scr-siRNA; scr-siRNA: Cells infected with lentivirus-mediated scramble small interfering RNA; NOB1-siRNA: Cells infected with lentivirus-mediated NOB1-siRNA.
- Citation: He XW, Feng T, Yin QL, Jian YW, Liu T. NOB1 is essential for the survival of RKO colorectal cancer cells. World J Gastroenterol 2015; 21(3): 868-877
- URL: https://www.wjgnet.com/1007-9327/full/v21/i3/868.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i3.868