Copyright
©The Author(s) 2015.
World J Gastroenterol. Jun 28, 2015; 21(24): 7375-7399
Published online Jun 28, 2015. doi: 10.3748/wjg.v21.i24.7375
Published online Jun 28, 2015. doi: 10.3748/wjg.v21.i24.7375
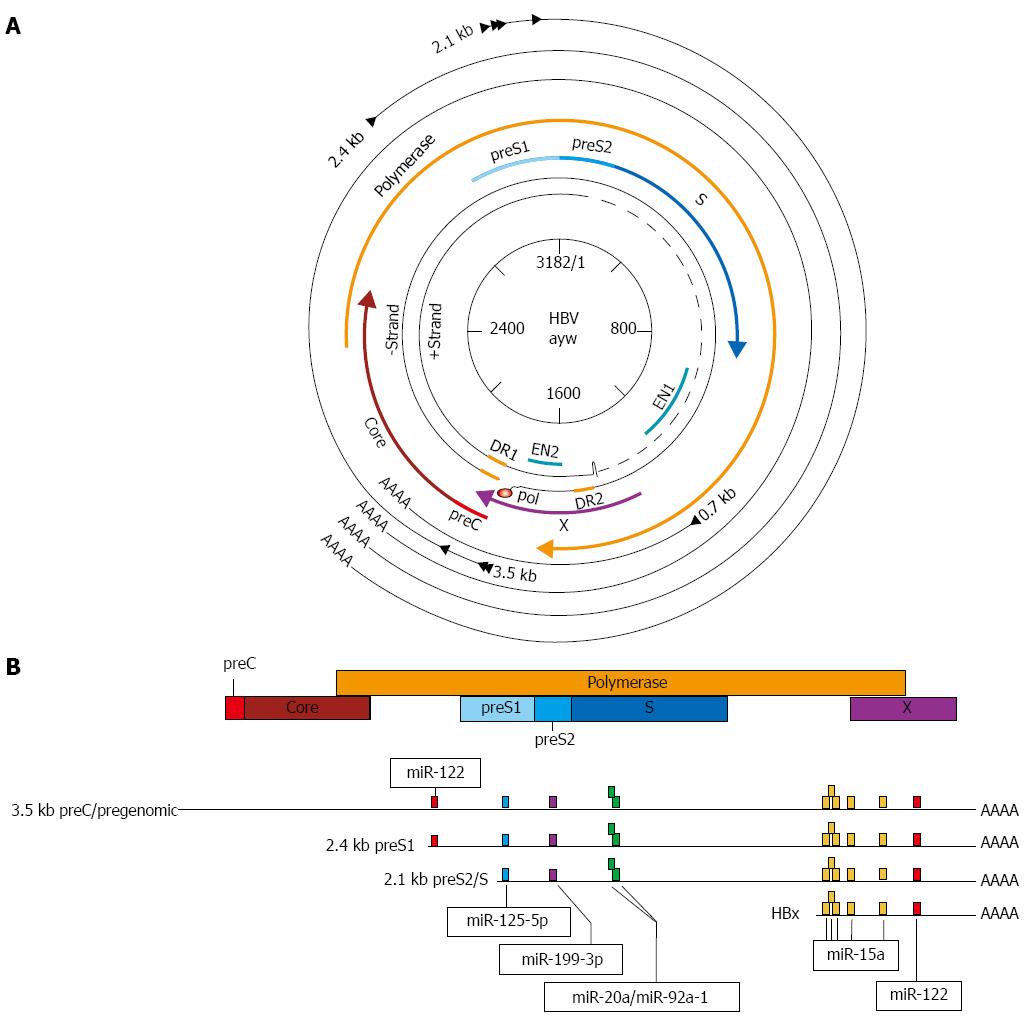
Figure 1 Genome structure of hepatitis B virus with proposed microRNA target sites.
A: The partially double-stranded DNA genome of hepatitis B virus (HBV) is depicted at the center, surrounded by the overlapping open-reading frames (ORFs) (thick colored arrows). The outer, black arrows represent the viral mRNA transcripts; B: Linear arrangement of overlapping ORFs and viral transcripts. Transcripts and ORF are aligned with each other according to genomic location and are to scale. Colored boxes on transcripts indicate proposed miRNA target sites discussed in the text.
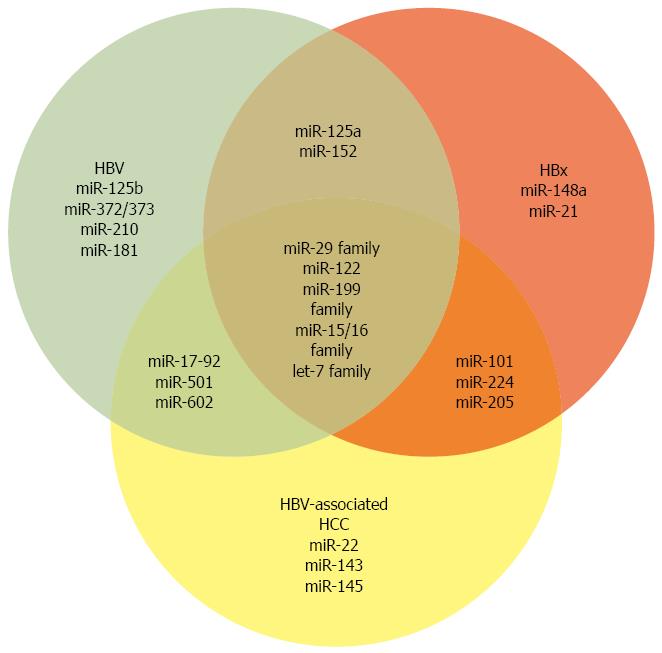
Figure 2 microRNAs altered by hepatitis B virus and -X protein or in hepatitis B virus-associated hepatocellular carcinoma.
Venn diagram depicting microRNAs discussed in the text that have altered expression levels in the context of hepatitis B virus (HBV) replication, HBV-X protein (HBx) expression, and/or HBV-associated hepatocellular carcinoma (HCC).
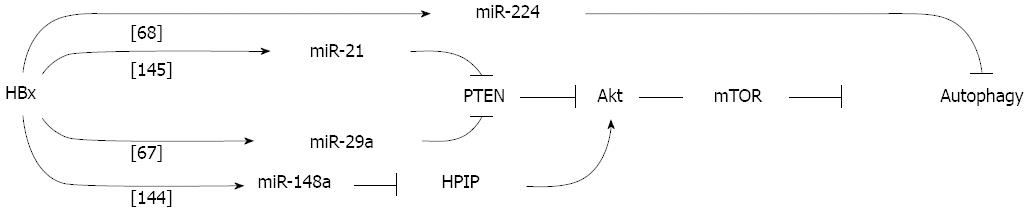
Figure 3 Model of potential hepatitis B virus X protein-mediated metabolic regulatory network.
Potential regulatory network involving hepatitis B virus (HBV)-X protein (HBx)-regulated microRNAs (miRNAs) and their downstream targets. See text for description and abbreviations.
- Citation: Lamontagne J, Steel LF, Bouchard MJ. Hepatitis B virus and microRNAs: Complex interactions affecting hepatitis B virus replication and hepatitis B virus-associated diseases. World J Gastroenterol 2015; 21(24): 7375-7399
- URL: https://www.wjgnet.com/1007-9327/full/v21/i24/7375.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i24.7375