Copyright
©2014 Baishideng Publishing Group Inc.
World J Gastroenterol. Dec 14, 2014; 20(46): 17498-17506
Published online Dec 14, 2014. doi: 10.3748/wjg.v20.i46.17498
Published online Dec 14, 2014. doi: 10.3748/wjg.v20.i46.17498
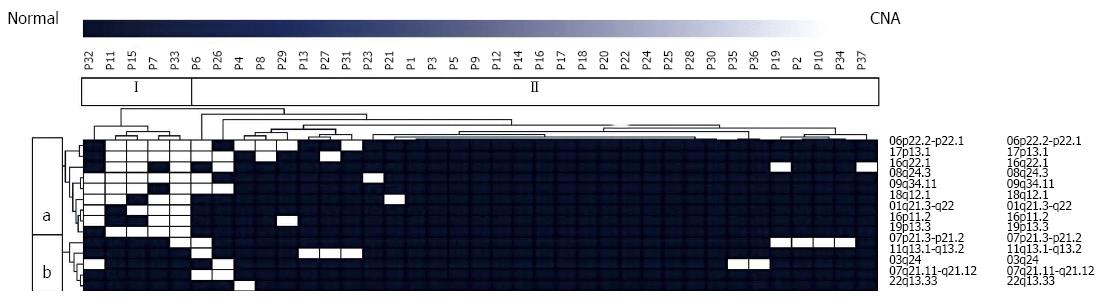
Figure 1 Genomic data of primary pancreatic neuroendocrine tumors obtained by array-based comparative genomic hybridization were subjected to unsupervised hierarchical cluster analysis utilizing Euclidean distance and average linkage in order to detect sub-groups of tumors exhibiting a significant overlap with regard to recurrently observed copy number alterations.
Two genomically distinct types of pancreatic neuroendocrine tumors were identified. Tumor group I included 5 tumors and group II consisted of 32 tumor samples. Moreover, we detected two distinct chromosomal clusters (a, b). Cluster a consisted of 9 loci recurrently harboring aberrations, whereas cluster b included 5 regions with frequent genomic imbalances. CNA: Copy number alterations.
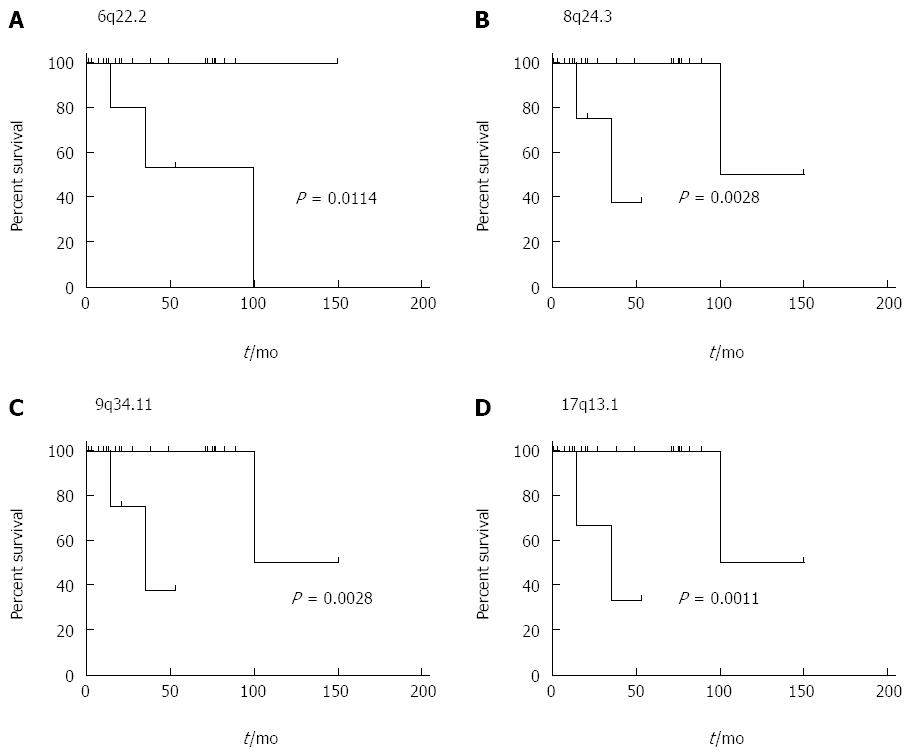
Figure 2 In order to assess overall survival and potential associations with recurrent chromosomal aberrations the log rank test was utilized and results were illustrated by Kaplan-Meier plots for both tumor groups as well as regions of recurrent genomic imbalance and patient survival employing the Mantel-Cox test.
Copy number alterations affecting 6p22.2-22.1 (A), 8q24.3 (B), 9q34.11 (C) and 17p13.1 (D) were shown to concur with significantly shortened overall survival.
- Citation: Gebauer N, Schmidt-Werthern C, Bernard V, Feller AC, Keck T, Begum N, Rades D, Lehnert H, Brabant G, Thorns C. Genomic landscape of pancreatic neuroendocrine tumors. World J Gastroenterol 2014; 20(46): 17498-17506
- URL: https://www.wjgnet.com/1007-9327/full/v20/i46/17498.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i46.17498