Copyright
©2014 Baishideng Publishing Group Inc.
World J Gastroenterol. Jul 7, 2014; 20(25): 7993-8004
Published online Jul 7, 2014. doi: 10.3748/wjg.v20.i25.7993
Published online Jul 7, 2014. doi: 10.3748/wjg.v20.i25.7993
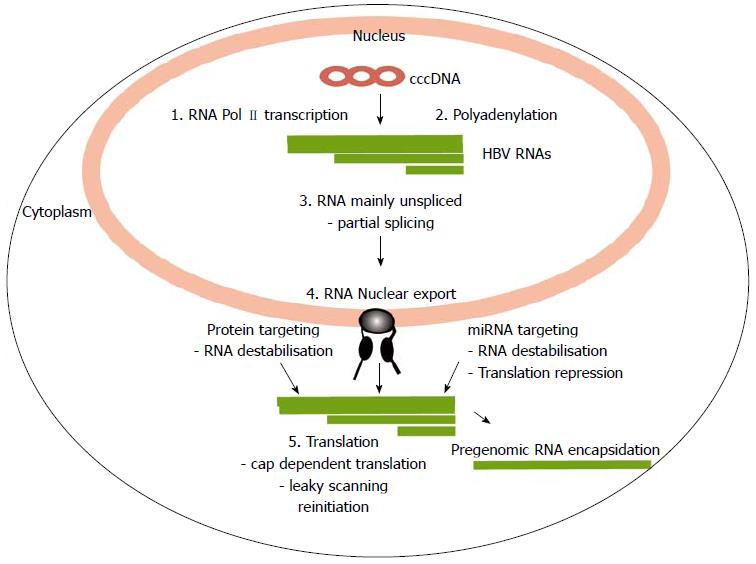
Figure 1 Overview of the steps of post-transcriptional processing and translation of hepatitis B virus RNAs.
The circular hepatitis B virus (HBV) genome is represented as the closed circular DNA (cccDNA). It encodes four large overlapping open reading frames encoding the core (C), surface (S), polymerase (P) and X proteins. It is transcribed into four main transcripts (HBV RNAs) with common polyadenylation sites and 3’ ends. Sites in which the virus uses processes that differ from the host are indicated and reviewed in the text (1) Transcription at specific viral promoters; (2) Non-canonical polyadenylation; (3) Partial splicing; (4) Nuclear export; and (5) Translation by leaky scanning and reinitiation.
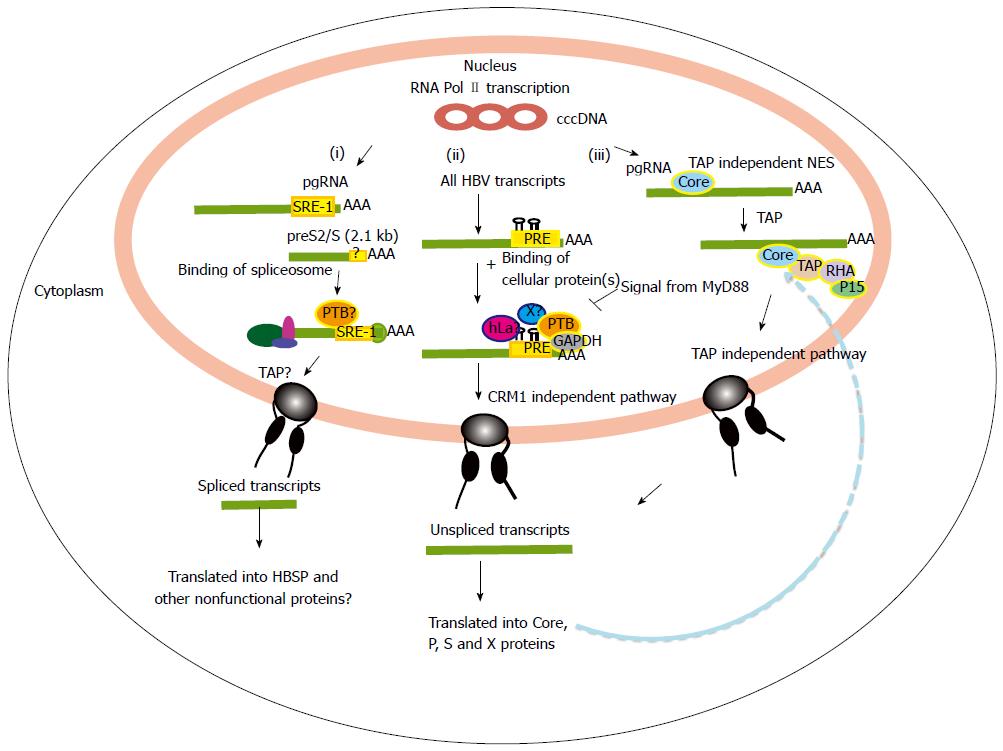
Figure 2 Nuclear export of hepatitis B virus mRNAs.
(i) Nuclear export for aberrant spliced hepatitis B virus (HBV) transcripts. In this pathway, HBV post-transcriptional regulatory element (PRE) enhances over-splicing of the pgRNA and perhaps also the S transcripts. The spliced RNAs are then possibly exported out via a TAP pathway. These spliced HBV transcripts are translated into apparently non-functional proteins such as HBSP; (ii) Nuclear export of major HBV unspliced and intronless transcripts e.g. the preS/S and X transcripts. For this pathway, HBV PRE plays roles in inhibiting splicing as well as mediating the nuclear export of the unspliced transcripts. The exact mechanism of this nuclear export pathway is unknown, but may involve binding of cellular proteins (PTB, GAPDH, hLa) near to conserved secondary structures of HBV PRE; and (iii) TAP dependent NES pathway of the pgRNA transcript. The core protein binds the pgRNA facilitating its export though the TAP/NFX1 pathway. SRE-1: Splicing regulatory element-1; NES: Nuclear export signals.
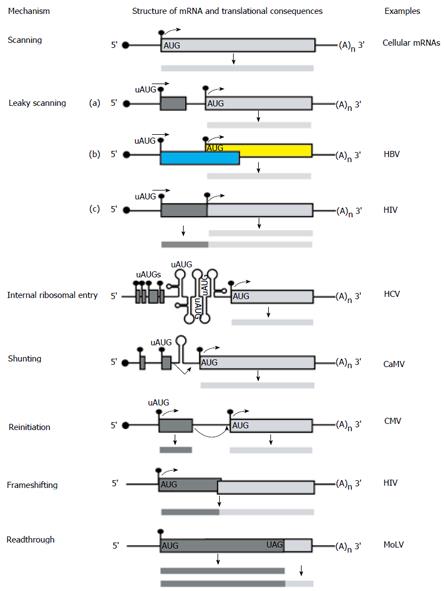
Figure 3 Alternative translation initiation mechanisms used by viruses and cellular mRNAs in eukaryotic cells.
The line with a filled circle represents a capped and polyadenylated mRNA with a 5′ UTR and open reading frame. The filled circle represents the cap, filled rectangle denotes an open reading frames, (A)n represents the polyA tail and the curved arrowhead denotes translation initiation. Various strategies for initiating protein synthesis on each mRNA and for decoding its information are outlined. The most common strategy used for cellular mRNAs is the scanning model. Other unconventional translational strategies employed by eukaryotes and exploited by infecting viruses included internal ribosomal entry, shunting, leaky scanning, reinitiation[54], frameshifting and readthrough[53]. Human immunodeficiency virus and CaMV are related to hepatitis B virus[55].
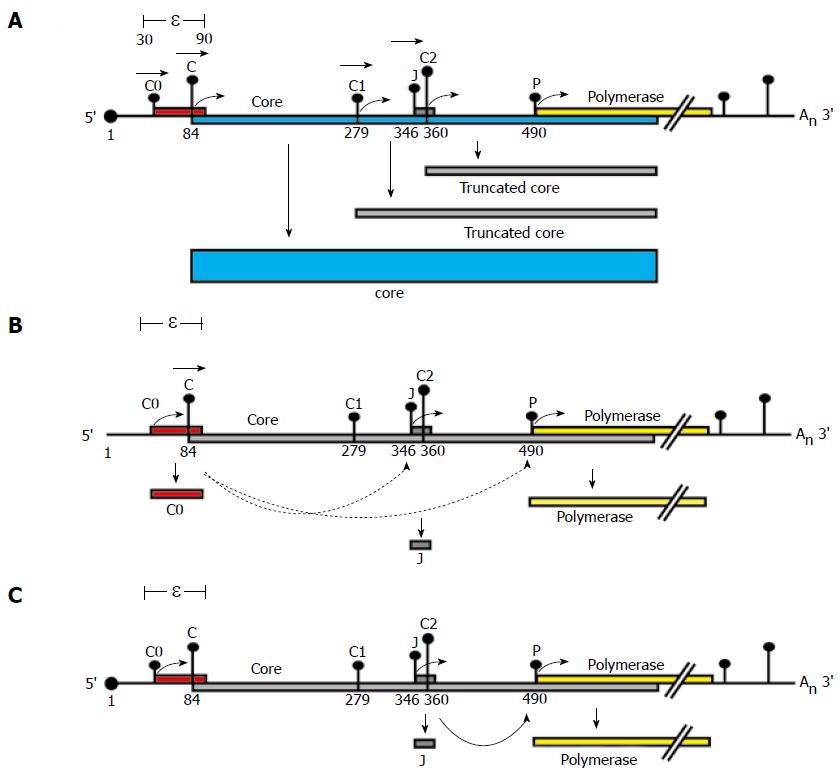
Figure 4 Proposed model for P protein synthesis.
A schematic representation of a pgRNA transcript and its coding regions. The pgRNA transcript encodes the core (blue) and polymerase (yellow) genes. The pgRNA transcript start site is numbered 1. The pgRNA contains the ε structure and a repeated sequence of 117 bases in both its 5′ and 3′ ends. Vertical bars with dots represent the initiation codons within the pgRNA labelled C0, C, C1, J, C2 and P. The height of each bar represents the match to the "ideal" initiation context (Kozak’s consensus)[15]. Filled boxes represent ORFs encoded by each initiation codon, (A)n denotes the polyA tail and the black dot at the 5′ end denotes the cap structure. The proposed model for P protein synthesis extends a scanning model. A: Leaky scanning. Ribosomes would scan from the capped 5′ end of the pgRNA, ribosomes would predominantly scan past C0 (in a poor initiation context) and initiate at the C AUG codon (which has an optimal initiation context) synthesising the core. However, some ribosomes may continue to leaky scan (represented by horizontal arrowheads) past the C AUG initiating at distal AUGs (e.g. making truncated core proteins from C1 or C2); B: Role of C0. The C0 initiation codon is the first in the pgRNA, some ribosomes would initiate here, when they terminate they could re-initiate at P or J (represented by lower curved arrows). Those that initiate at P would translate the polymerase; C: Role of J. Ribosomes which bypass C and C1 AUG codons by leaky scanning would then initiate translation at the J AUG codon (optimal initiation context), translating the short J ORF effectively bypassing the C2 AUG codon. Ribosomes which translated the J ORF may then reinitiate at the P AUG codon, translating the polymerase (curved arrow).
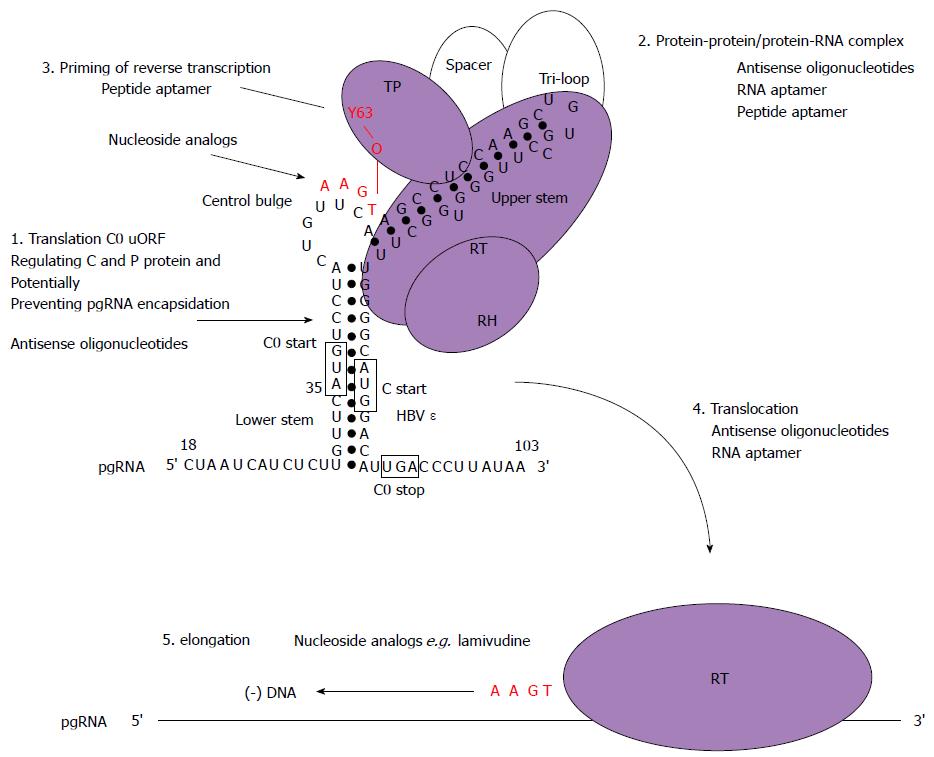
Figure 5 Schematic model for the initiation of reverse transcription in hepatitis B virus with stages targetable by existing drugs, as well as potential targets for antiviral compounds.
These may serve as a basis to design new strategies combining drugs with different mechanisms of action. Domains of the P protein are abbreviated as follows: terminal protein (TP), RNase H domain (RH), reverse transcriptase domain (RT). Open circles represent cellular chaperones. The initiation of reverse transcription to generate a dsDNA genome involves the pgRNA which is also the template for C and P protein translation. In this model, the C0 ORF which spans the ε structure is also translated (albeit weakly) and could potentially discriminate the selection of pgRNA required for encapsidation and translation (step 1). Secondly, the hepatitis B virus (HBV) P protein would then interact with cellular chaperones (heat shock proteins), the ε structure and gain enzymatic activity (step 2). Next, the reverse transcription step is primed by the formation of covalent bond between conserved tyrosine of the TP domain and the first nt of the minus-strand DNA followed by additions of three more nucleotides, GAA in a template dependent manner (step 3). Following an arrest in the viral DNA synthesis, a strand transfer occurs and the complex of P protein and short DNA primer is translocated to the 3′ end of the template DR1 (step 4). Then, elongation of the minus-strand DNA continues (step 5). Currently, the triphosphate forms of nucleoside analogs have shown inhibitory effects on the priming reaction (targeting step 3) or interfere with viral minus-strand DNA elongation (targeting step 5).
- Citation: Chen A, T-Thienprasert NP, Brown CM. Prospects for inhibiting the post-transcriptional regulation of gene expression in hepatitis B virus. World J Gastroenterol 2014; 20(25): 7993-8004
- URL: https://www.wjgnet.com/1007-9327/full/v20/i25/7993.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i25.7993