Copyright
©2014 Baishideng Publishing Group Inc.
World J Gastroenterol. Jun 21, 2014; 20(23): 7104-7122
Published online Jun 21, 2014. doi: 10.3748/wjg.v20.i23.7104
Published online Jun 21, 2014. doi: 10.3748/wjg.v20.i23.7104
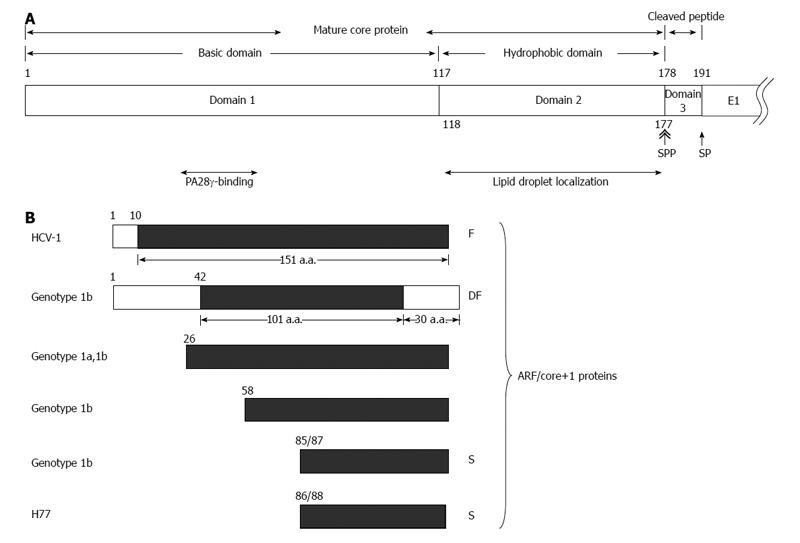
Figure 1 Various hepatitis C virus core gene products.
A: The hepatitis C virus (HCV) polyprotein is cleaved at residues 191/192 by the host signal peptidase (SP) and further cleaved at residue 177/178 by signal peptide peptidase (SPP) to release the mature core protein (a.a. 1-177) and the cleaved peptide (a.a. 178-191) from the precursor polyprotein. The mature core protein consists of the positively charged domain 1 (a.a. 1-117) and the hydrophobic domain 2 (a.a. 118-177). The highly basic domain 1 is involved in RNA-binding and its oligomerization. The region containing residues 44-71 of domain 1 binds to PA28γ. Domain 2 is involved in the association of HCV core protein with lipid droplets; B: Different alternative reading frame (ARF)/core+1 proteins from different HCV isolates/genotypes. The polypeptides from the conventional open reading frame are marked by empty rectangles while those from the alternative reading frame (ARF/core+1) by filled rectangles. The termination codon of ARF/core+1 proteins from other isolates/genotypes may be different from those shown in this figure.
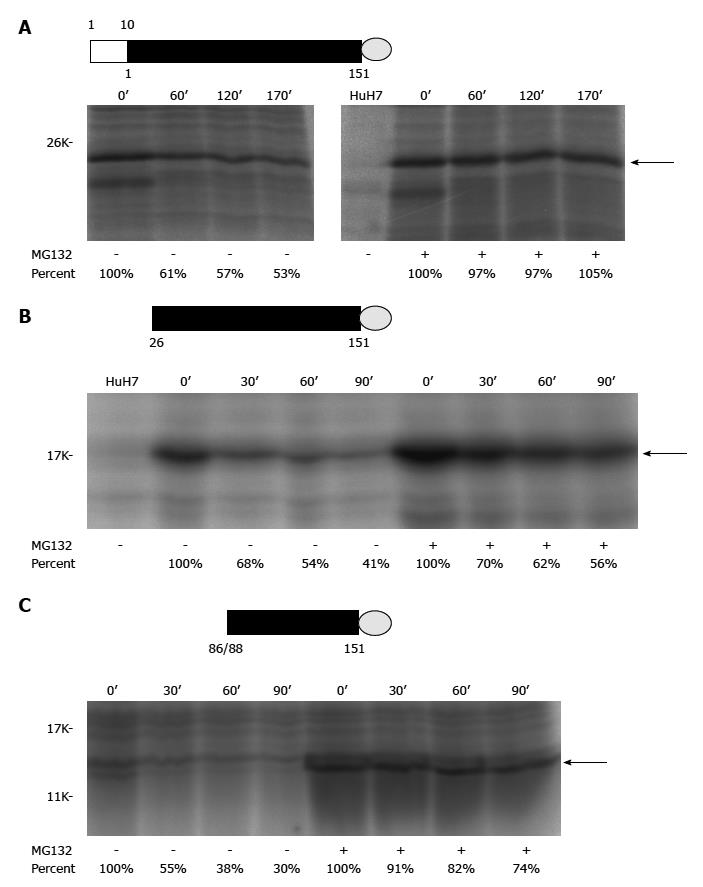
Figure 2 Pulse-chase experiments of different recombinant alternative reading frame/core+1 proteins (H77 sequence was used for this study).
A: Authentic F[alternative reading frame (ARF)/core+1/F] protein (the empty rectangle marks the sequence overlapping with the core protein while the filled rectangle for the core+1 coding sequence) with a V5 tag (represented by a gray circle) at its C-terminus; B: The ARF/core+1 protein translated from AUG of amino acid 26 with a V5 tag at its C-terminus; C: The ARF/core+1 protein translated from AUG of amino acids 86/88 with a V5 tag at its C-terminus. HuH7 cells were mock-transfected or transfected with various constructs expressing different recombinant ARF/core+1 proteins as indicated. Forty-eight hours after transfection, cells were incubated in methionine-free medium for two hours and subsequently radiolabeled with 35S-methionine in the same medium (160 mCi/mL) for two hours. Then, regular medium with or without MG132 treatment was used for further cultivation. At the indicated times, cells were disrupted and proteins were extracted to perform the immunoprecipitation assay using rabbit anti-F polyclonal antibody.
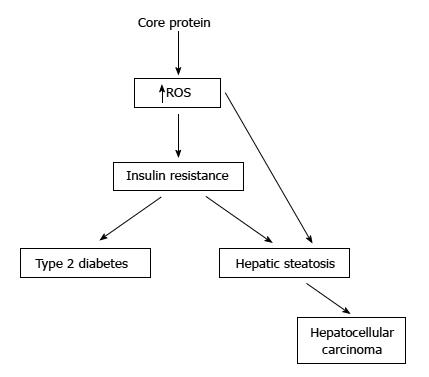
Figure 3 Pathogenecity of hepatitis C virus core protein in the transgenic mice.
Some studies have showed that the transgenic mice with core protein developed steatosis only, or steatosis followed by hepatocellular carcinoma. In other studies, the transgenic mice with constitutive core protein expression developed insulin resistance, then leading to type 2 diabetes on a high-fat diet. Most of these mice would develop hepatic steatosis and some of them would even develop hepatocellular carcinoma.
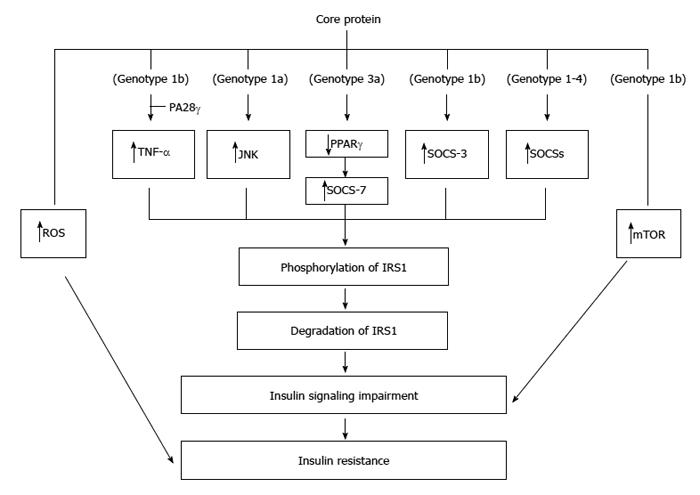
Figure 4 Molecular mechanisms regarding the insulin resistance induced by core protein.
Core proteins from different isolates/genotypes may use common and/or distinct mechanisms to cause insulin resistance.
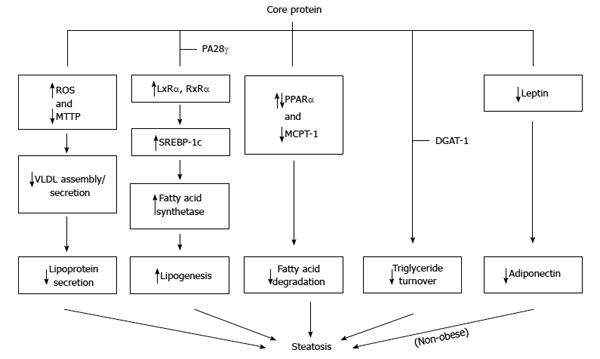
Figure 5 Molecular mechanisms proposed for the steatosis caused by core protein.
Core protein from one genotype may use more than one mechanism to induce steatosis.
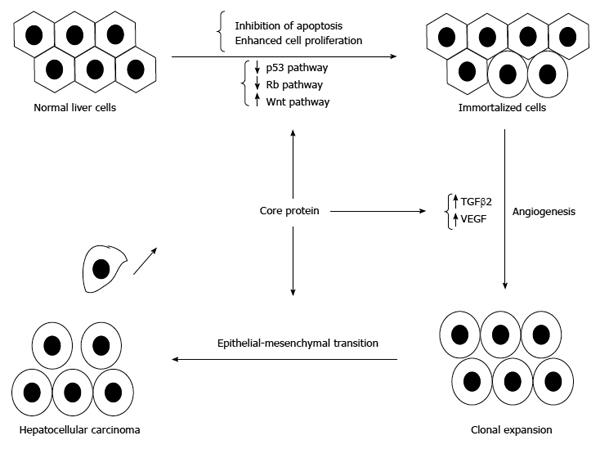
Figure 6 Involvement of core protein in the development of hepatocellular carcinoma.
In addition to inducing immortalization of hepatocytes, core protein could enhance angiogenesis and promote the epithelial-mesenchymal transition to result in hepatocellular carcinoma.
- Citation: Li HC, Ma HC, Yang CH, Lo SY. Production and pathogenicity of hepatitis C virus core gene products. World J Gastroenterol 2014; 20(23): 7104-7122
- URL: https://www.wjgnet.com/1007-9327/full/v20/i23/7104.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i23.7104