Copyright
©2014 Baishideng Publishing Group Co.
World J Gastroenterol. Mar 21, 2014; 20(11): 2785-2800
Published online Mar 21, 2014. doi: 10.3748/wjg.v20.i11.2785
Published online Mar 21, 2014. doi: 10.3748/wjg.v20.i11.2785
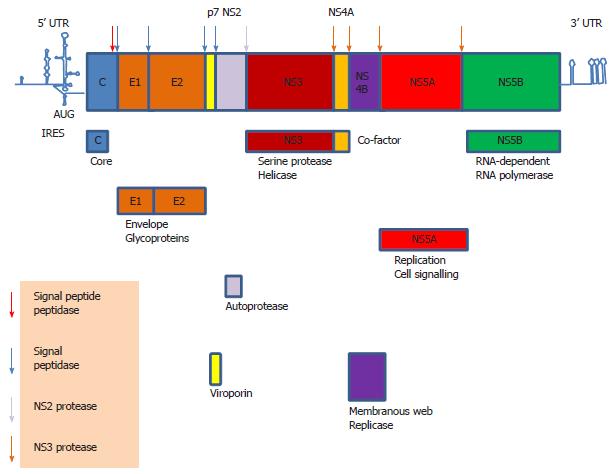
Figure 1 Hepatitis C virus genome.
The 9.6 kb hepatitis C virus genome is flanked by highly structured 5’ untranslated region (UTR) and 3’ UTR which contain regulatory elements. The genomic sequence is translated into a polyprotein via internal ribosome entry site (IRES)-mediated translation. The polyprotein is then cleaved into individual structural and non-structural (NS) proteins by cellular signal peptidase and signal peptide peptidase and viral auto protease (NS2) and serine protease (NS3).
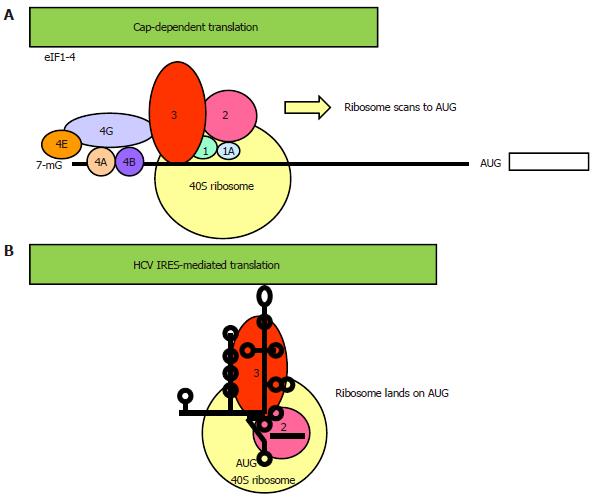
Figure 2 Two different modes of translational initiation.
A: Cap-dependent translation. Initiation of translation of most cellular genes requires a 7-methyl guanosine cap-structure at its 5’ end and a full set of canonical eukaryotic translation factors (eIFs) for ribosome binding and scanning to the AUG start codon[29]. The cap-binding complex eIF4F consists of the cap-binding eIF4E, the scaffold eIF4G and the helicase eIF4A. eIF4B promotes the RNA unwinding activity of eIF4A. Recruitment of the 40S ribosomal subunit-eIF3-eIF2 pre-initiation complex to the 5’ end of the mRNA is via protein-protein interaction between eIF4G and eIF3. The ribosomal complex, primed by eIF1/1A, then scans a short distance (50-100 nucleotides) to (usually) the nearest AUG triplet to initiate translation; B: Internal ribosome entry site (IRES)-mediated translation. The viral IRES element spans a considerably longer 5’ untranslated region (UTR) (180-450 nucleotides) that folds into a higher order structure and is interspersed with multiple AUG triplets[37]. The requirement for canonical eIFs varies greatly amongst IRESs, ranging from the dependence of the entire set of eIFs in the picornavirus hepatitis A virus IRES to none of them in the cricket paralysis virus IRES. The hepatitis C virus (HCV) IRES has the second simplest requirement, only needing eIF2 and eIF3. In reminiscent of the prokaryotic ribosomal binding of the Shine-Dalgarno sequence, the HCV IRES can directly recruit the 40S ribosome which lands on the authentic AUG initiator codon[35,63,65]. In some other IRESs, the ribosome lands on an upstream AUG and then scans or shunts to the initiator AUG[37]. An important feature about IRES-mediated translation is that its activity is regulated by IRES trans-acting factors[43].
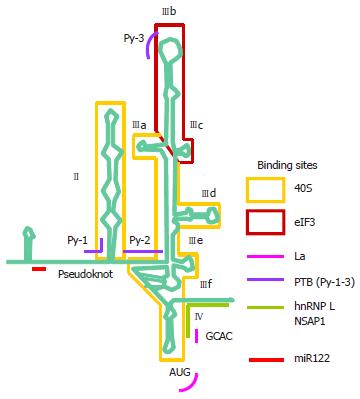
Figure 3 The hepatitis C virus internal ribosome entry site.
The hepatitis C virus (HCV) 5’ untranslated region (UTR) is divided into four stem-loop domains and sub-domains[61]. The internal ribosome entry site (IRES) element is made up of approximately 340 nucleotides and spans domain II and extends into the core-coding region encompassing a double pseudoknot fold[13,62-64]. The stem, loop and unpaired regions are all important in interaction with host factors. The HCV IRES can directly recruit the 40S ribosomal subunit before association with eukaryotic translation factor (eIF)2 and eIF3 to form the 43S pre-initiation complex[35,65]. Structural and biochemical studies have indicated an unusually vast binding site for the ribosome encompassing domains II, III and IV and conformational changes in both the 40S ribosomal subunit and the IRES have been observed upon their interaction[66]. Binding between the IRES and the ribosome is thought to be mediated via ribosomal proteins although the role of RNA-RNA interaction cannot be excluded[67-69]. eIF3 can bind both the ribosome and junction domain IIIabc and domain IIIb. The ternary complex eIF2α-guanosine triphosphate (GTP)-the tRNA for the first methionine (MettRNAi) does not directly bind the IRES, rather it forms a ternary complex with eIF3 and the 40S ribosomal subunit to position MettRNAi directly onto the AUG codon in the P-site of the ribosome[70]. A number of non-canonical host factors are able to bind the HCV IRES and likely play a facilitating role in IRES translation. However, there is evidence for a critical role of the La autoantigen in IRES translation, by binding to and altering the conformation of the IRES to orchestrate assembly of the ribosomal complex[82,83]. NS: Non-structural.
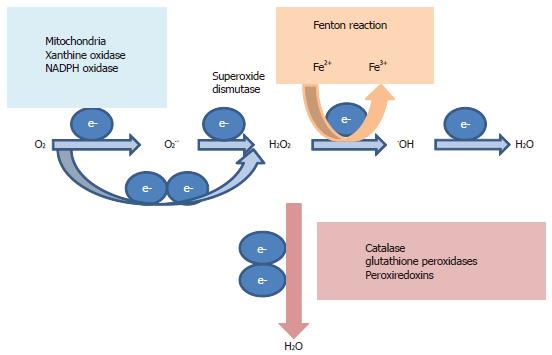
Figure 4 Oxidant and anti-oxidant systems.
During active respiration in the mitochondria, electron leaked from the respiratory chain reacts with oxygen (O2) to form reactive oxygen species (ROS) superoxide anion O2.- and pro-oxidant-hydrogen peroxide (H2O2). O2.- is quickly dismuted to H2O2 in a reaction catalyzed by superoxide dismutase. Non-mitochondrial sources of O2.- and H2O2 include cytosolic xanthine oxidase and plasma membrane nicotinamide adenine dinucleotide phosphate (NADPH) oxidase. H2O2 is decomposed by iron in the Fenton reaction to yield the highly oxidizing hydroxyl radical .OH, causing macromolecule damage, DNA damage and lipid peroxidation. H2O2 can be reduced to water (H2O) by catalase, glutathione peroxidases and peroxiredoxins.
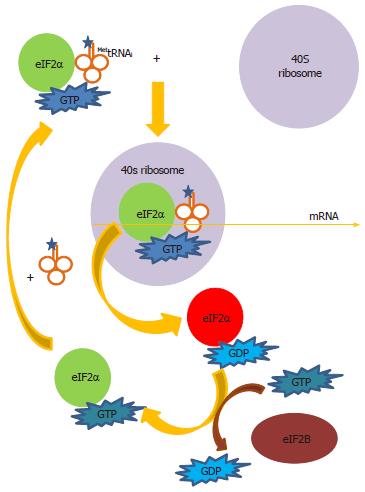
Figure 5 Delivery of MettRNAi to the ribosome.
Eukaryotic translation factor (eIF)2α forms a ternary complex with guanosine triphosphate (GTP) and the tRNA for the first methionine (MettRNAi) to deliver MettRNAi to the 40S ribosomal subunit. Following hydrolysis of GTP to guanosine diphosphate (GDP), the eIF2α-GDP complex leaves the ribosome. GDP is converted to GTP in an exchange reaction catalyzed by the exchanger eIF2B. eIF2α-GTP can then be recycled for more ternary complex formation with MettRNAi to continue translation initiation. When eIF2α is phosphorylated at Serine-51 (red), it inhibits eIF2B. Thus GDP cannot be exchanged for GTP and protein synthesis is arrested at this step.
- Citation: Chan SW. Establishment of chronic hepatitis C virus infection: Translational evasion of oxidative defence. World J Gastroenterol 2014; 20(11): 2785-2800
- URL: https://www.wjgnet.com/1007-9327/full/v20/i11/2785.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i11.2785