Copyright
©2013 Baishideng Publishing Group Co.
World J Gastroenterol. Nov 7, 2013; 19(41): 6995-7023
Published online Nov 7, 2013. doi: 10.3748/wjg.v19.i41.6995
Published online Nov 7, 2013. doi: 10.3748/wjg.v19.i41.6995
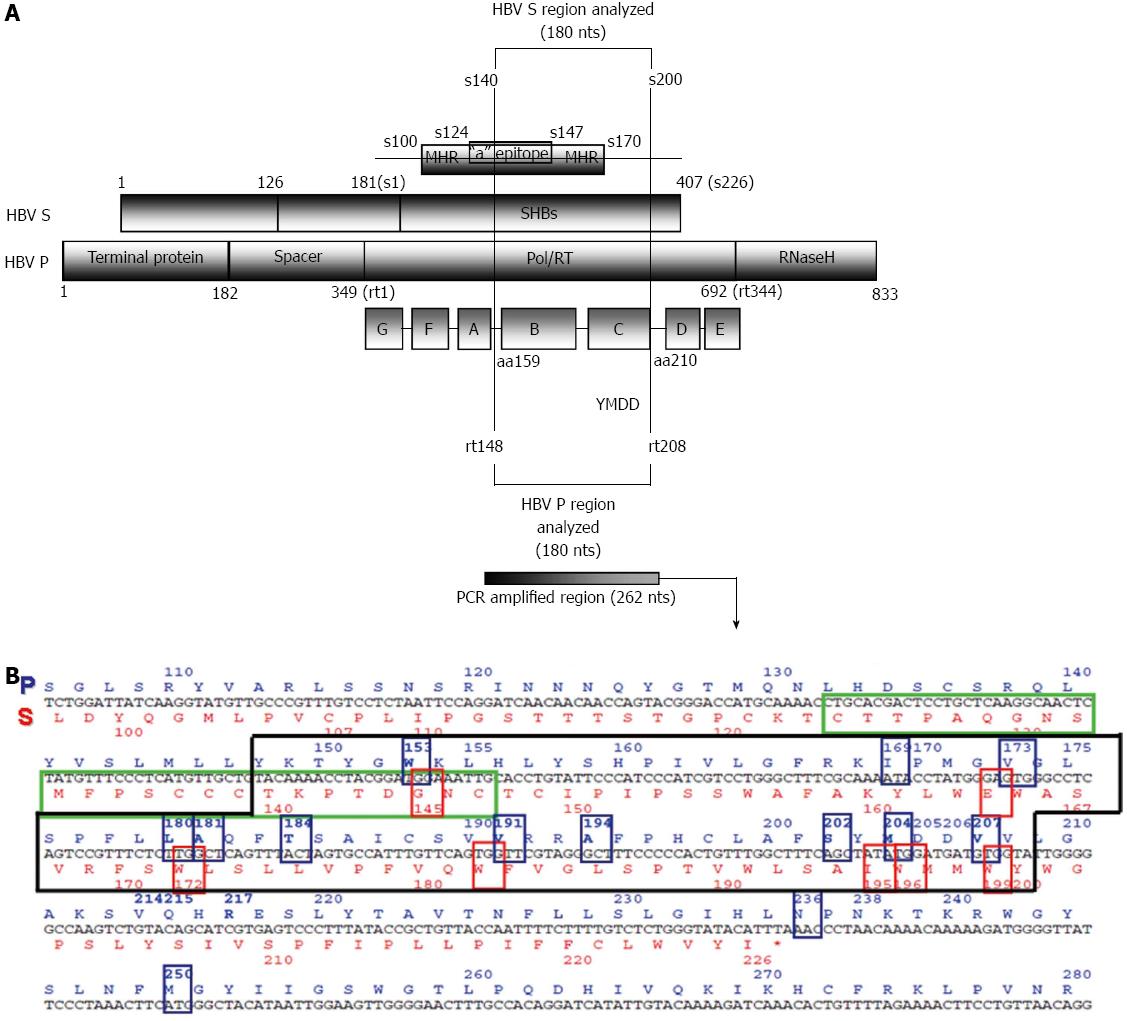
Figure 1 Overlapping region of the polymerase and surface genes (A) and its corresponding sequence (B).
Translation into amino acids is depicted in blue in the P open reading frame (ORF) (above) and red in the S ORF (below). Most of the main codons related to nucleos(t)ide resistance (framed in blue), and the overlapping codons in the S ORF that may give rise to immune escape or stop codons (framed in red) are located within the fragment analyzed in a previous study by our group[58].
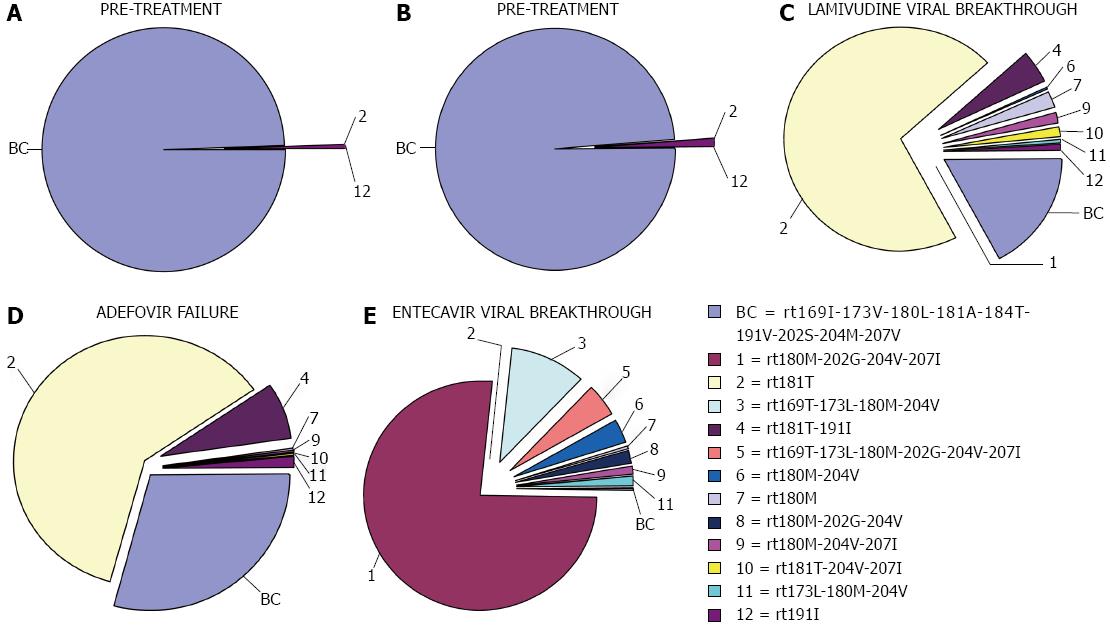
Figure 2 Changes in percentages of reverse transcriptase variants during follow-up of a patient included in one of our studies.
Reproduced from Rodriguez-Frías et al[58]. BC: Baseline combination.
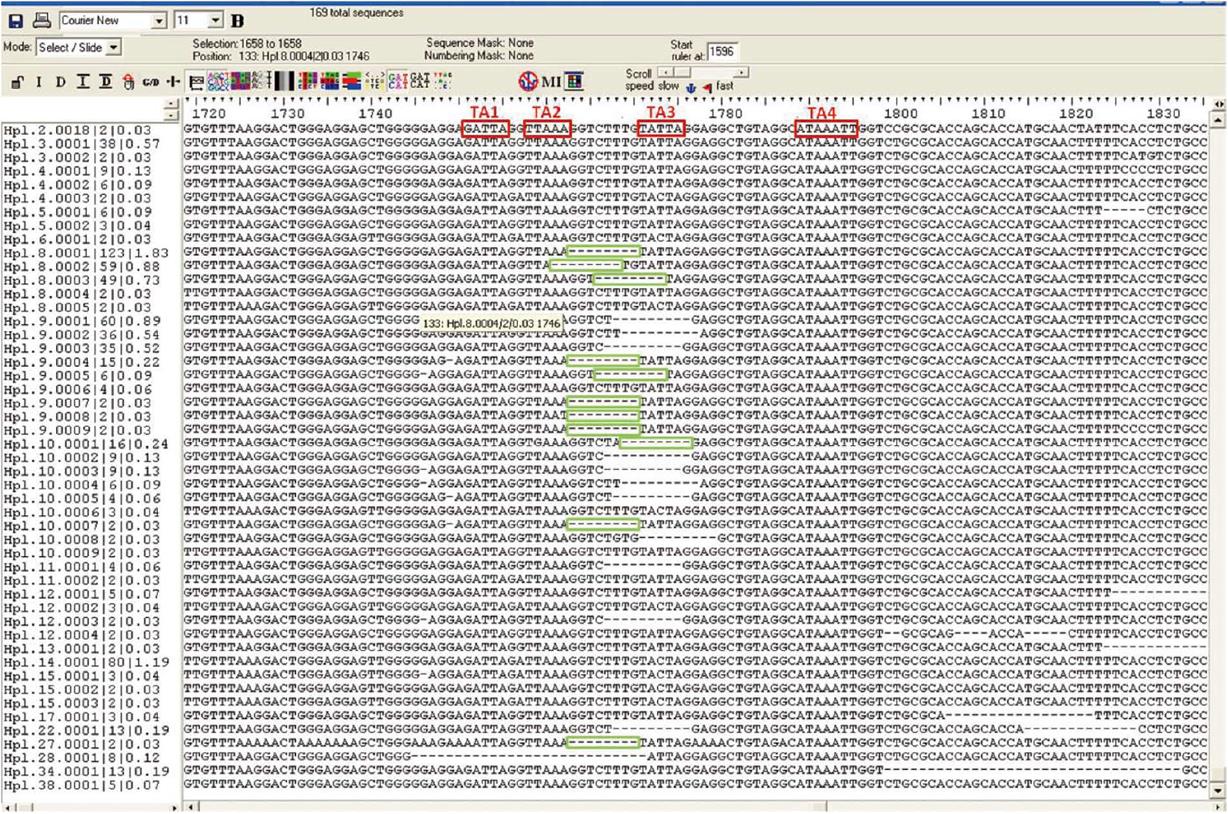
Figure 3 Quasispecies of the X gene obtained by unpublished result.
The four TA-like boxes are highlighted in red and the eight nucleotides detected are indicated in green.
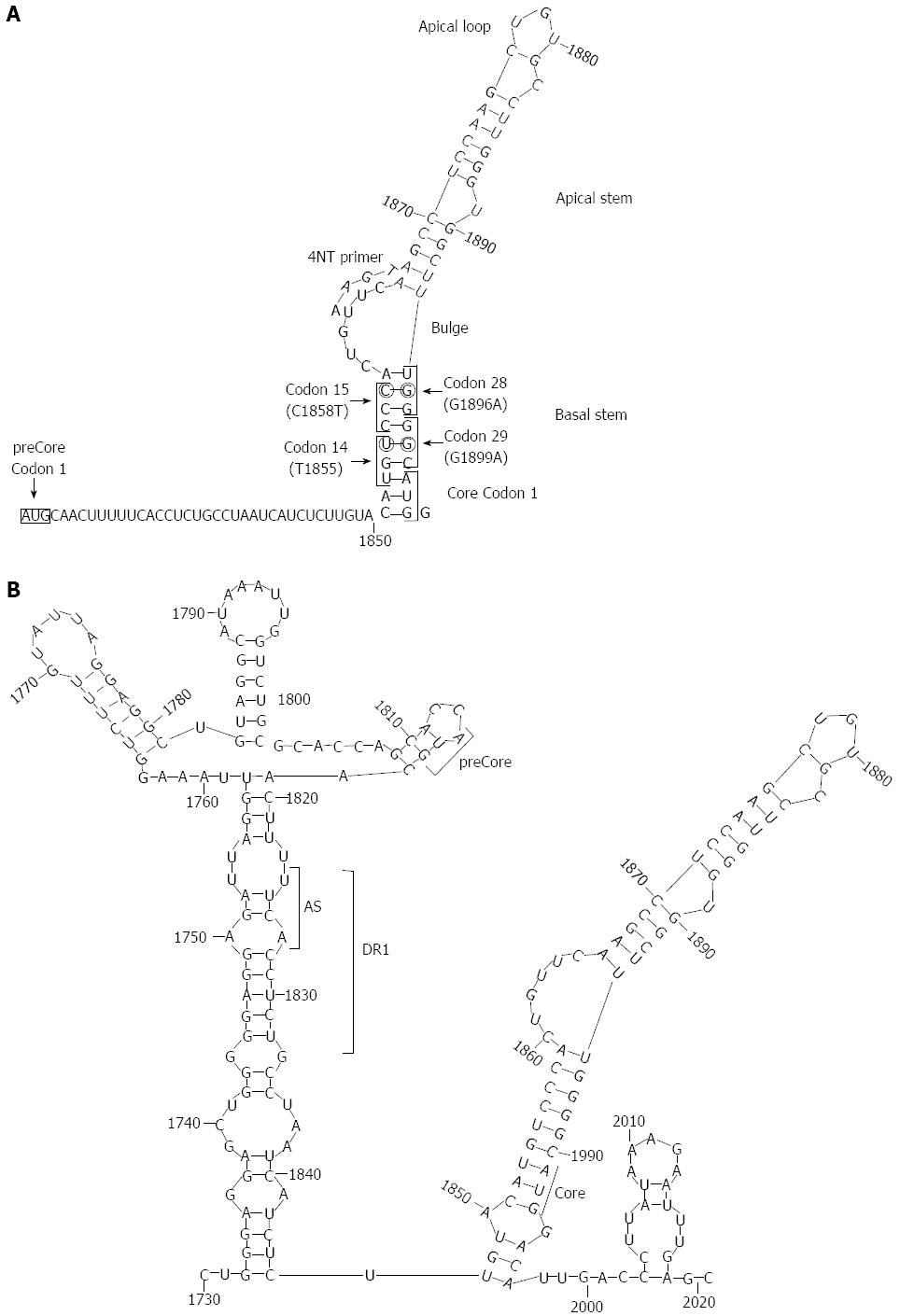
Figure 4 Secondary structures adopted at the 5’ and 3’ ends of hepatitis B virus pgRNA reported by our group.
A: Stem-loop structure of the 5’ with main structural motifs (loops and stems), main codons (1, 14, 15, 28, and 29 of preCore, and 1 of Core gene), and the location of 4-nt primer annealing in the 5’; B: Stem-loop structure of the 3’. The acceptor site (AS), direct repeat region 1 (DR1), and preCore and Core start codons are indicated in the figure. Reproduced from Homs et al[56].
- Citation: Rodriguez-Frias F, Buti M, Tabernero D, Homs M. Quasispecies structure, cornerstone of hepatitis B virus infection: Mass sequencing approach. World J Gastroenterol 2013; 19(41): 6995-7023
- URL: https://www.wjgnet.com/1007-9327/full/v19/i41/6995.htm
- DOI: https://dx.doi.org/10.3748/wjg.v19.i41.6995