Copyright
©2008 The WJG Press and Baishideng.
World J Gastroenterol. May 21, 2008; 14(19): 3006-3014
Published online May 21, 2008. doi: 10.3748/wjg.14.3006
Published online May 21, 2008. doi: 10.3748/wjg.14.3006
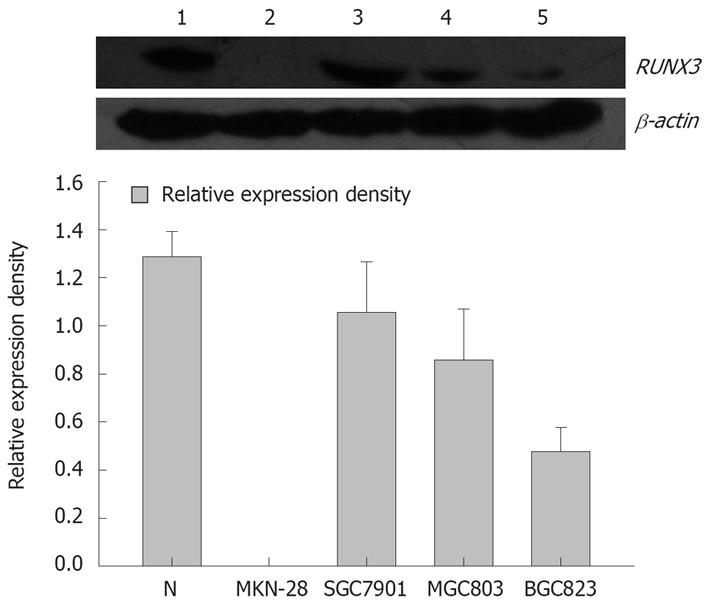
Figure 1 RUNX3 expression in the gastric cancer cells detected by Western blot.
1: Positive control: Normal gastric tissue; 2: MKN-28 cells; 3: SGC-7901 cells; 4: MGC-803 cells; 5: BGC-823 cells.
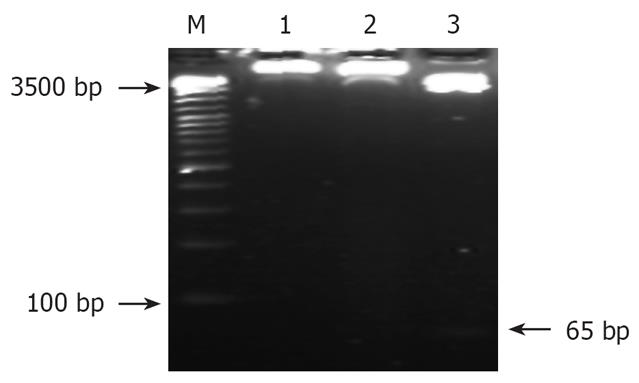
Figure 2 Identification of recombinant by restrict endonucleases digestion.
M: DNA Ladder; 1: pSilencer3.1-H1/SGC7901; 2: pSilencer3.1-H1-shRNA/RUNX3/SGC-7901; 3: pSilencer3.1-H1-shRNA/RUNX3/SGC7901 cut with BamHI/HindIII.
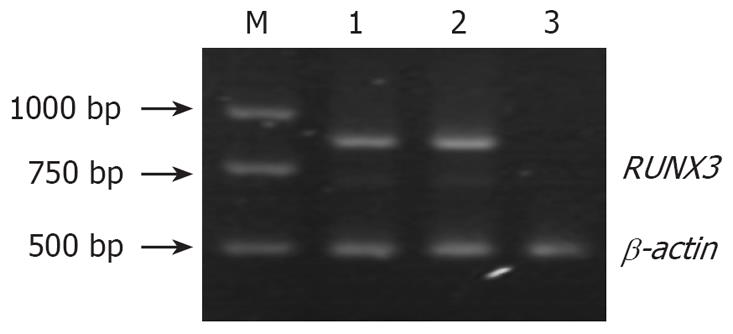
Figure 3 RUNX3 expression in the three groups of cells detected by RT-PCR.
M: DL2000 marker; 1: SGC7901 cells; 2: pSilencer3.1-H1 /SGC7901 cells cells; 3: pSilencer3.1-H1-shRNA/RUNX3/SGC7901 cells.
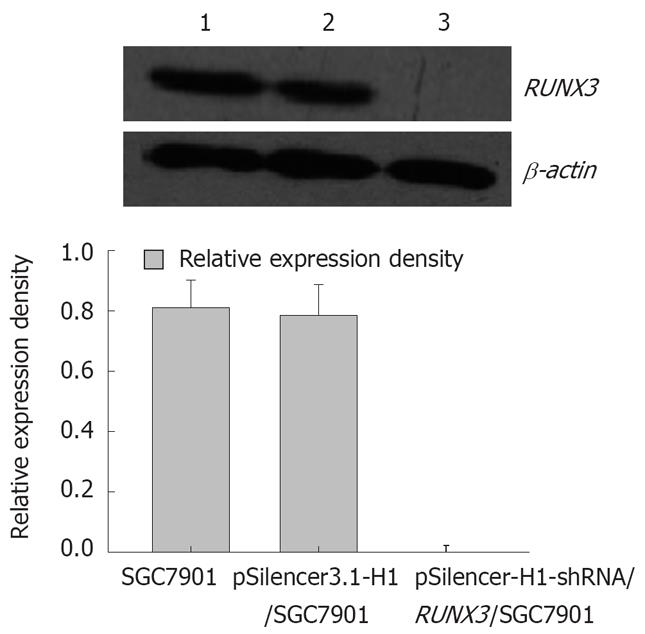
Figure 4 RUNX3 expression detected by Western blot.
1: SGC7901 cells; 2: pSilencer3.1-H1/c SGC7901 cells; 3: pSilencer3.1-H1-shRNA/RUNX3/SGC7901 cells.
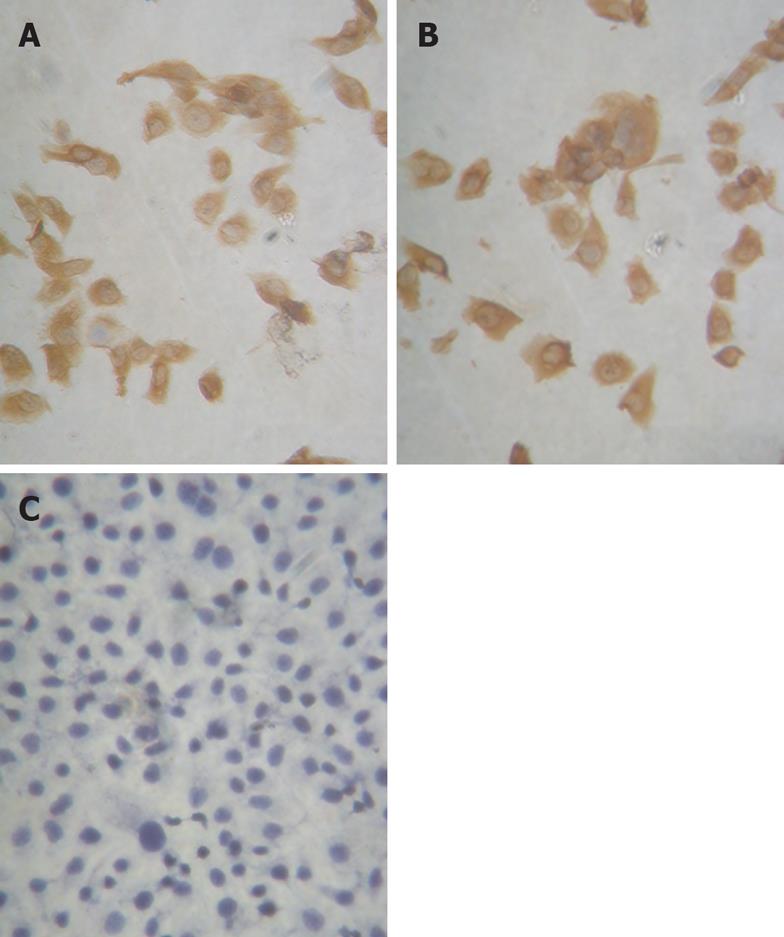
Figure 5 RUNX3 expression in SGC7901 cells by immunohistochemistry (× 400).
A: SGC7901 cells, the protein of RUNX3 expressed and located in the endochylema and the nucleus in the SGC7901 cell without transfection; B: pSilencer3.1-H1/SGC7901 cells, the protein of RUNX3 expressed and located in the endochylema and the nucleus in the SGC7901 cell with the plasmid DNA of pSilencer3.1-H1; C: pSilencer3.1-H1-shRNA/RUNX3/SGC7901 cells, the protein of RUNX3 lost expression in the SGC790 cells transfected with the recombinated plasmid-pSilencer 3.1-H1-shRNA/RUNX3.
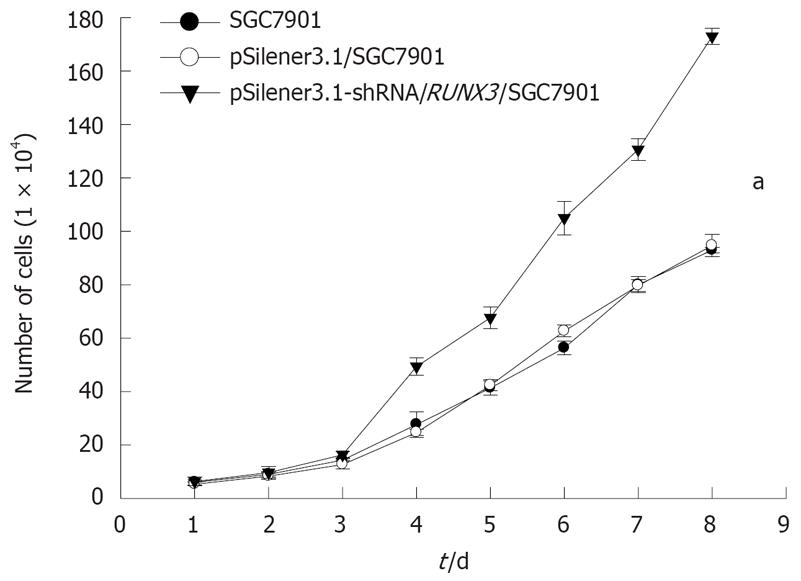
Figure 6 The growth effect of SGC7901 cell by silencing RUNX3 (aP < 0.
05).
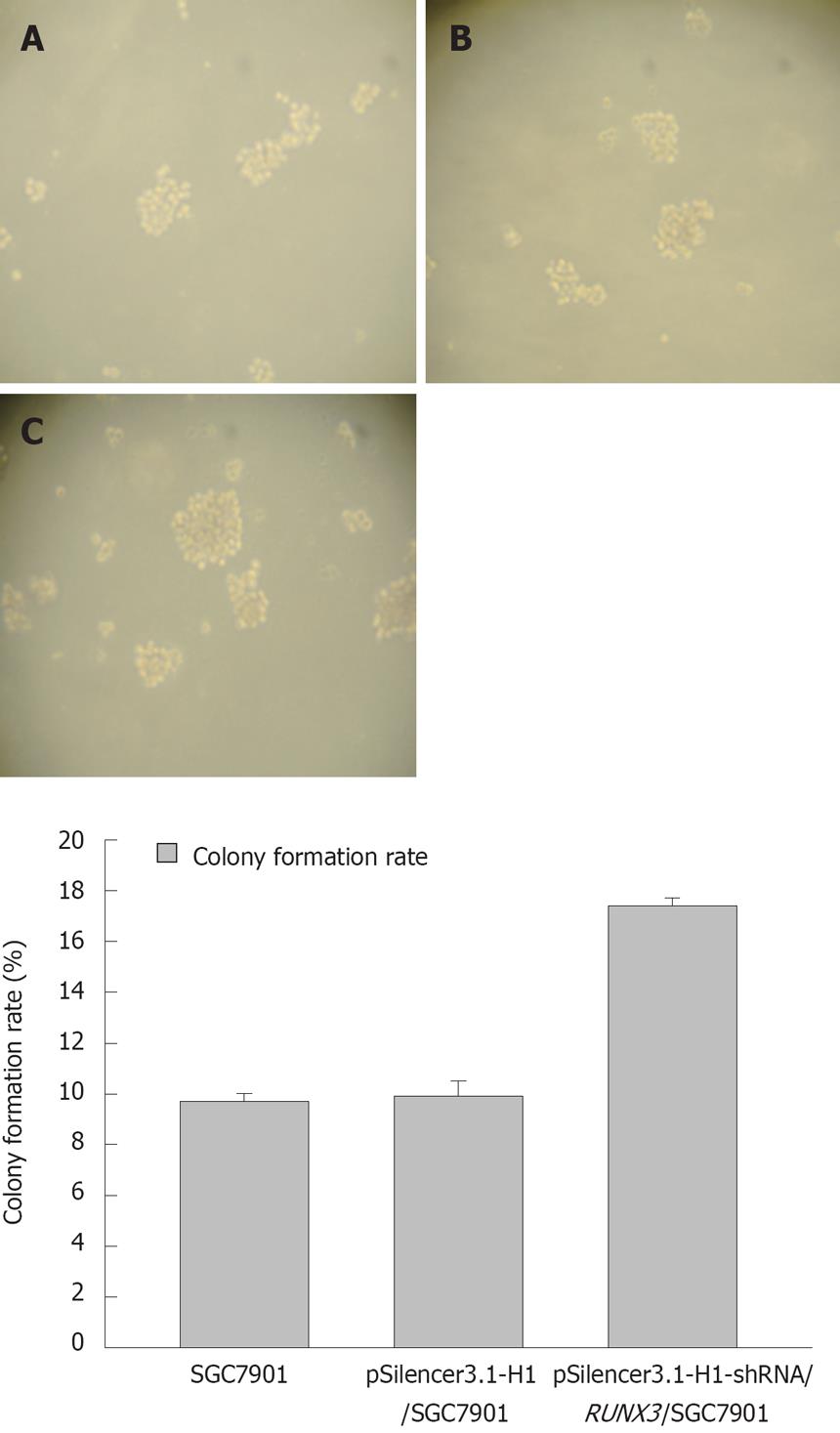
Figure 7 The colony formation assay of SGC7901 in the soft agar (×100).
A: SGC7901 cells, the cloning efficiency was 9.9% ± 0.3% in the SGC7901 cell without transfection; B: pSilencer3.1-H1/SGC7901 cells, the cloning efficiency was 9.7% ± 0.6% in the SGC7901 cell with the plasmid DNA of pSilencer3.1-H1; C: pSilencer3.1-H1-shRNA/RUNX3/SGC 7901 cells, the cloning efficiency was 17.4% ± 0.31% in the SGC790 cells transfected with the recombinated plasmid-pSilencer3.1-H1-shRNA/RUNX3. A significant increase of the colony formation rate in the pSilencer3.1-H1-shRNA/RUNX3/SGC790 cells was discovered compared with the controls-the SGC-79011cells and pSilencer3.1-H1/SGC7901cells (P < 0.01).
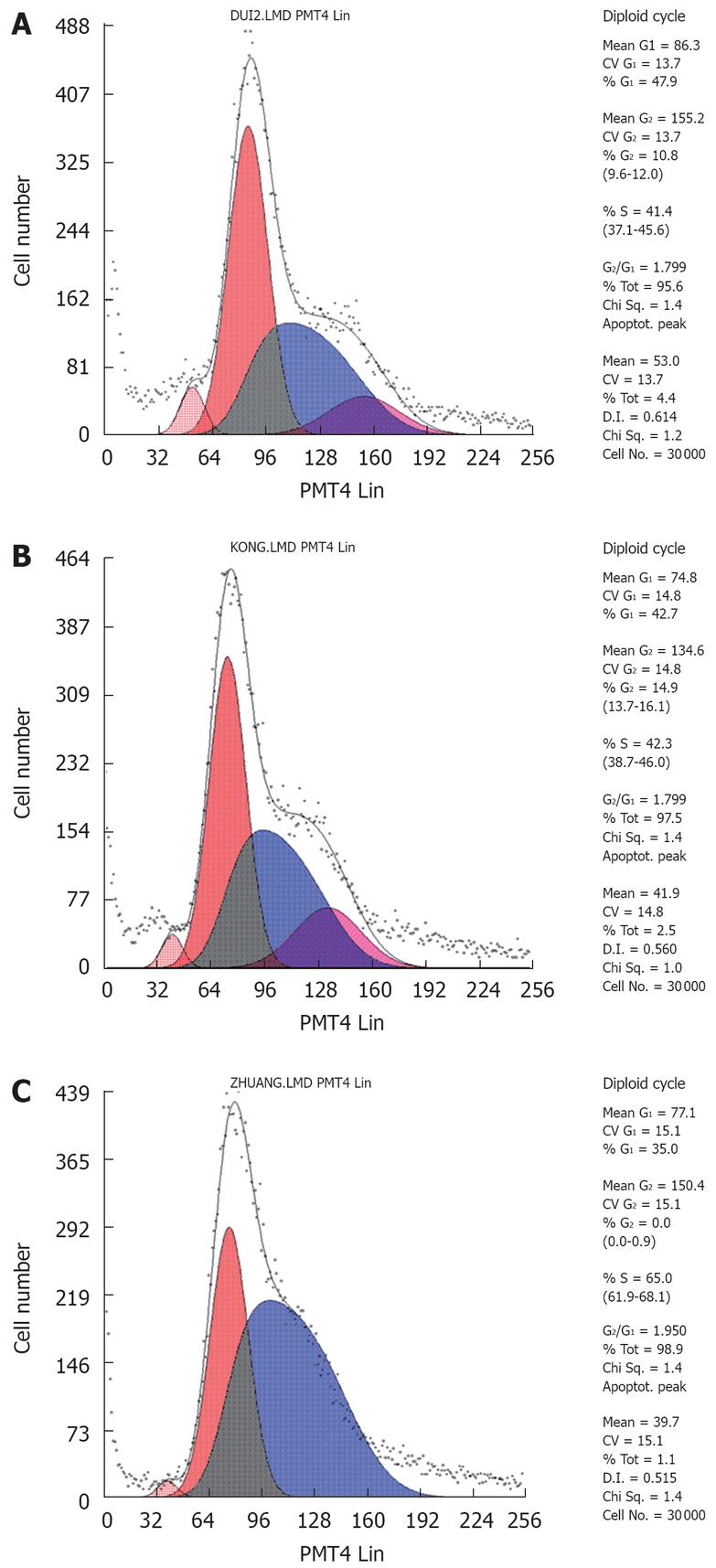
Figure 8 The cell cycle analysis by FCM.
A: SGC790 cells, the cell proportions of G0/G1 and S stages were 43.2% ± 1.2% and 47.7% ± 1.1% in the SGC7901cell without transfection, respectively; B: pSilencer3.1-H1/SGC7901 cells, the cell proportions of G0/G1 and S stages were 40.3% ± 2.0% and 49.3% ± 0.9% in the SGC7901 cell with the plasmid DNA of pSilencer3.1-H1 respectively; C: pSilencer3.1-shRNA/RUNX3SGC7901 cells, the cell proportions of G0/G1 and S stages were 37.2% ± 1.9% and 60.5% ± 0.8% in the SGC790 cells transfected with the recombinated plasmid-pSilencer3.1-H1-shRNA/RUNX3 respectively. Compared with that of G0/G1 and S stages of two controls, the cell proportions of G0/G1 and S stages in the pSilencer3.1-H1-shRNA/RUNX3/SGC7901 cells decreased and increased obviously, respectively (P < 0.05).
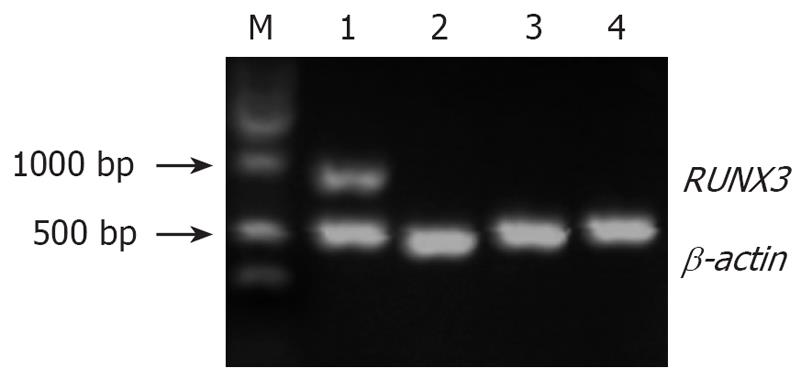
Figure 9 RUNX3 expresion detected by RT-PCR in the four groups of SGC7901 cells.
M: Marker; 1: Positive control, SGC7901 cells untreated with 5-Aza-CdR; 2: Negative control, pSilencer3.1-H1-shRNA/RUNX3/SGC7901cells untreated with 5-Aza-CdR; 3: Experimental group 1, pSilencer3.1-H1-shRNA/RUNX3/SGC7901cells treated with 5 × 10-6 mol/L 5-Aza-CdR; 4: Experimental group 2, pSilencer3.1-H1-shRNA/RUNX3/SGC7901 cells treated with 1 x 10-5mol/L 5-Aza-CdR.
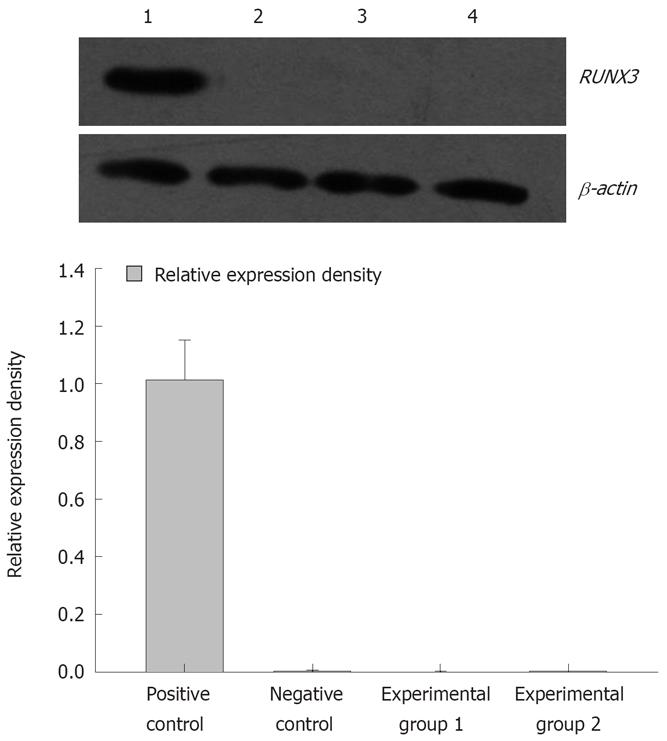
Figure 10 RUNX3 protein detected by Western blotting in the four groups of SGC7901 cells.
1: Positive control, SGC7901 cells untreated with 5-Aza-CdR; 2: Negative control, pSilencer3.1-H1-shRNA/RUNX3/SGC7901cells untreated with 5-Aza-CdR; 3: Experimental group 1, pSilencer3.1-H1-shRNA/RUNX3/SGC7901cells treated with 5 × 10-6 mol/L 5-Aza-CdR; 4: Experimental group 2, pSilencer3.1-H1-shRNA/RUNX3/SGC7901 cells treated with 1 × 10-5 mol/L 5-Aza-CdR.
-
Citation: Feng XZ, He XS, Zhuang YZ, Luo Q, Jiang JH, Yang S, Tang XF, Liu JL, Chen T. Investigation of transcriptional gene silencing and mechanism induced by shRNAs targeted to
RUNX3 in vitro . World J Gastroenterol 2008; 14(19): 3006-3014 - URL: https://www.wjgnet.com/1007-9327/full/v14/i19/3006.htm
- DOI: https://dx.doi.org/10.3748/wjg.14.3006