Copyright
©2007 Baishideng Publishing Group Co.
World J Gastroenterol. Feb 28, 2007; 13(8): 1195-1203
Published online Feb 28, 2007. doi: 10.3748/wjg.v13.i8.1195
Published online Feb 28, 2007. doi: 10.3748/wjg.v13.i8.1195
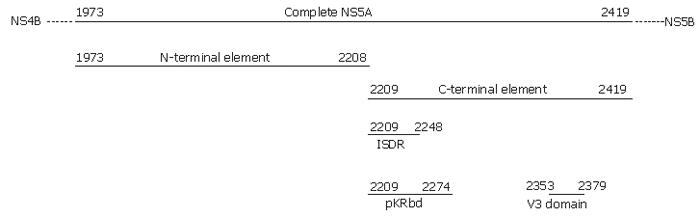
Figure 1 Schematic localisation of the six studied regions in NS5A protein.
Amino acids positions are numbered according to the HCV-J polyprotein and the NS5A region is within positions 1973-2419.
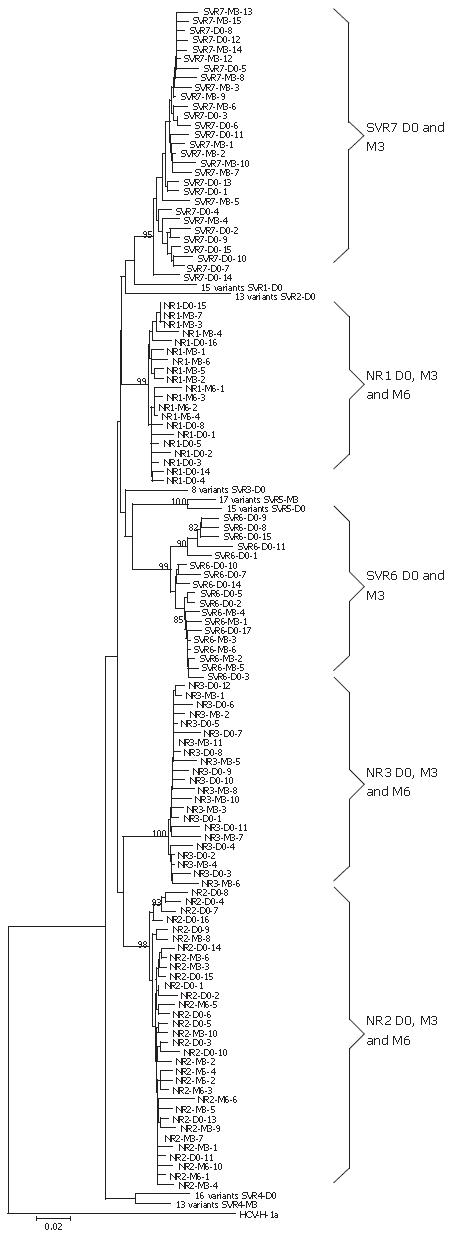
Figure 2 Phylogenetic tree of NS5A region (codon 1973-2419) of amino acids HCV variants obtained from ten patients at different time of treatment.
D0: beginning of the treatment; M3: 3 mo of treatment; SVR: Sustained Virological Responders to IFN-ribavirin therapy; NR: Non-Responders to IFN-ribavirin therapy. HCV-H strain was used as outgroup. The internal node numbers represent the bootstrap values. Distance scale represents 2% of difference between sequences. When all NS5A sequences within a patient at one time point of treatment are grouped in a cluster, this cluster is presented by only one node.
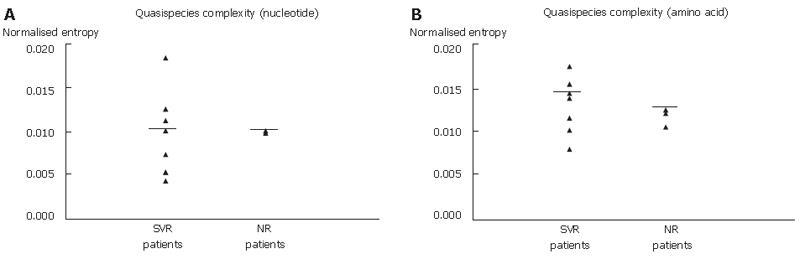
Figure 3 Genetic complexity of HCV quasispecies (A: nucleotide; B: amino acid) calculated by normalized entropy in seven sustained virological responders and three non-responders with median indicated by horizontal bar for each group.
P-value calculated by Mann-Whitney U-test.
- Citation: Veillon P, Payan C, Le Guillou-Guillemette H, Gaudy C, Lunel F. Quasispecies evolution in NS5A region of hepatitis C virus genotype 1b during interferon or combined interferon-ribavirin therapy. World J Gastroenterol 2007; 13(8): 1195-1203
- URL: https://www.wjgnet.com/1007-9327/full/v13/i8/1195.htm
- DOI: https://dx.doi.org/10.3748/wjg.v13.i8.1195