Copyright
©2007 Baishideng Publishing Group Co.
World J Gastroenterol. May 7, 2007; 13(17): 2416-2426
Published online May 7, 2007. doi: 10.3748/wjg.v13.i17.2416
Published online May 7, 2007. doi: 10.3748/wjg.v13.i17.2416
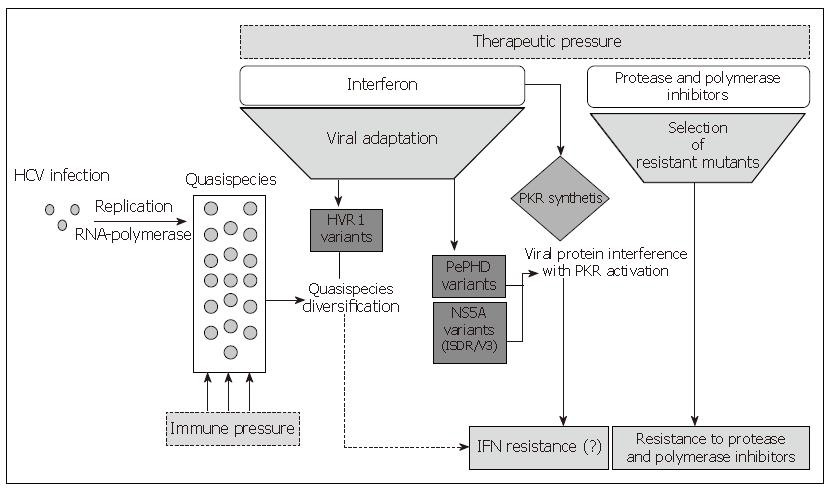
Figure 1 Mechanisms involved in HCV resistance to antiviral therapies.
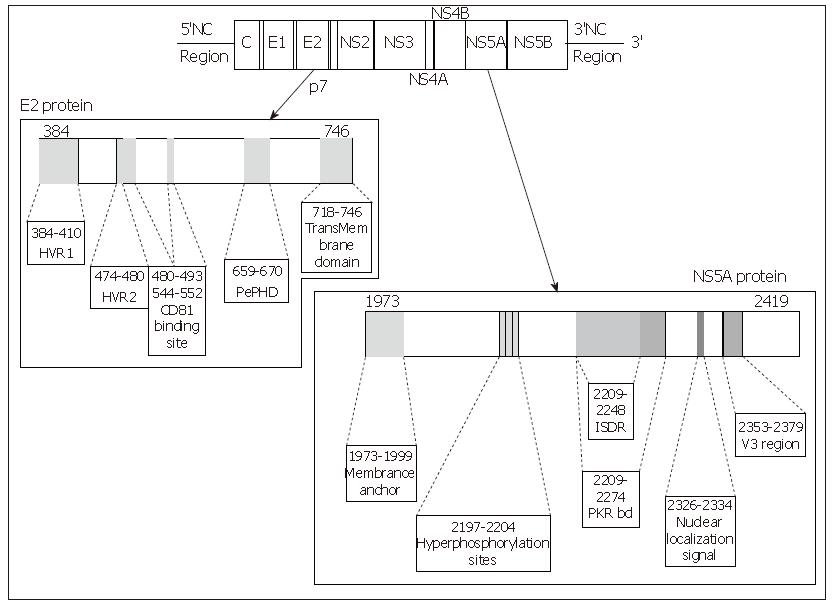
Figure 2 Linear representation of the E2 and NS5A proteins of the HCV, location of the main interesting domains of each protein.
Each site of interest is represented by a grey zone on the protein and is linked to a box containing its name and amino acid positions.
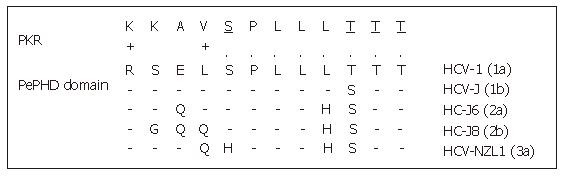
Figure 3 Sequence homology between HCV E2-PePHD domain and PKR.
PePHD domains from various HCV genotypes (1a, 1b, 2a, 2b, 3a) are aligned and compared with the PKR sequence. Identical and similar amino acids to PKR are shown by (.) and (+), respectively. PKR auto-phosphorylation sites are underlined.
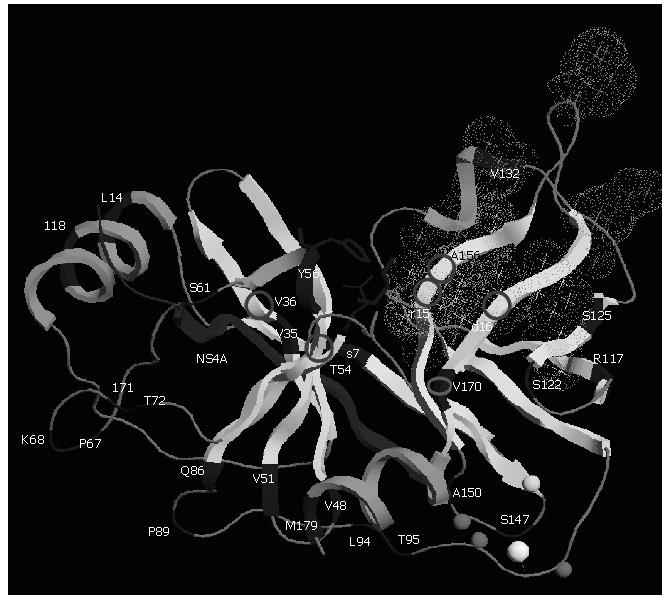
Figure 4 Three-dimensional structure of NS3 protease (PDB accession code 1NS3).
Polymorphism and main residues implicated in resistance are shown. The protease is shown based on its secondary structure in light grey. Main polymorphic residues are shown in dark grey. The side chains of the residues forming the catalytic triad (H57, D81, and S139) are displayed in dark grey ball-and-stick representation. The NS4A cofactor is shown in dark grey and Zinc ion as a white ball. Zn2+ ligands (C97, C99 and C149) and H145 residue are modeled in light grey spheres. Stars correspond to the side chains of the residues forming the S1 to S6 substrate binding pockets (grey dots cloud) of the enzyme. Residues implicated in resistance to protease NS3 inhibitors are shown with circles. This figure was prepared using the RasTop programme version 2.0.3-VF.
Figure 5 Three-dimensional structure of NS5B RNA-dependent RNA polymerase (PDB accession code 1QUV).
The three subdomains of the polymerase are shown (Fingers, Thumb and Palm) as well as the main residues targeted by each class of polymerase inhibitors (NNI 1: Non nucleoside class 1, NNI 2: Non nucleoside class 2, NNI 3: Non nucleoside class 3, NI: Nucleoside Inhibitor). This figure was prepared using the RasTop programme version 2.0.3-VF.
- Citation: Le Guillou-Guillemette H, Vallet S, Gaudy-Graffin C, Payan C, Pivert A, Goudeau A, Lunel-Fabiani F. Genetic diversity of the hepatitis C virus: Impact and issues in the antiviral therapy. World J Gastroenterol 2007; 13(17): 2416-2426
- URL: https://www.wjgnet.com/1007-9327/full/v13/i17/2416.htm
- DOI: https://dx.doi.org/10.3748/wjg.v13.i17.2416