Copyright
©2007 Baishideng Publishing Group Co.
World J Gastroenterol. May 7, 2007; 13(17): 2406-2415
Published online May 7, 2007. doi: 10.3748/wjg.v13.i17.2406
Published online May 7, 2007. doi: 10.3748/wjg.v13.i17.2406
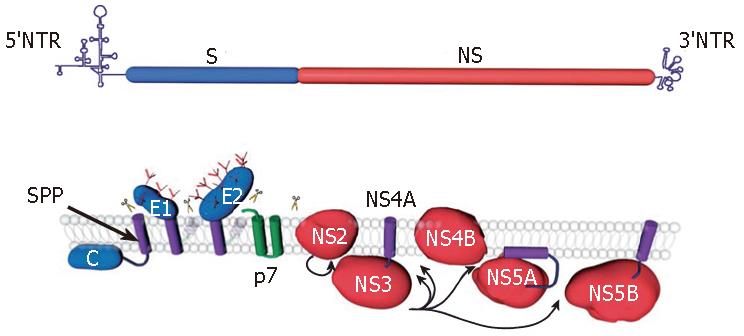
Figure 1 HCV genome organization (top) and polyprotein processing (bottom).
HCV encodes a single polyprotein with the structural proteins (S) and the nonstructural proteins (NS) present in the N-terminal one-third and the C-terminal two-third of the polyprotein, respectively. The polyprotein processing and the location of the 10 proteins relative to the endoplasmic reticulum membrane are schematically represented. Scissors indicate cleavages by a host signal peptidase. Arrows indicate NS2-3 and NS3-4A cleavages. The intramembrane arrow indicates cleavage by a host signal peptide peptidase (SPP). The transmembrane domains of E1 and E2 are shown after signal-peptidase cleavage and reorientation of their C-terminus. In addition, the precleavage topology of the transmembrane domains of E1 and E2 is shown in light grey.
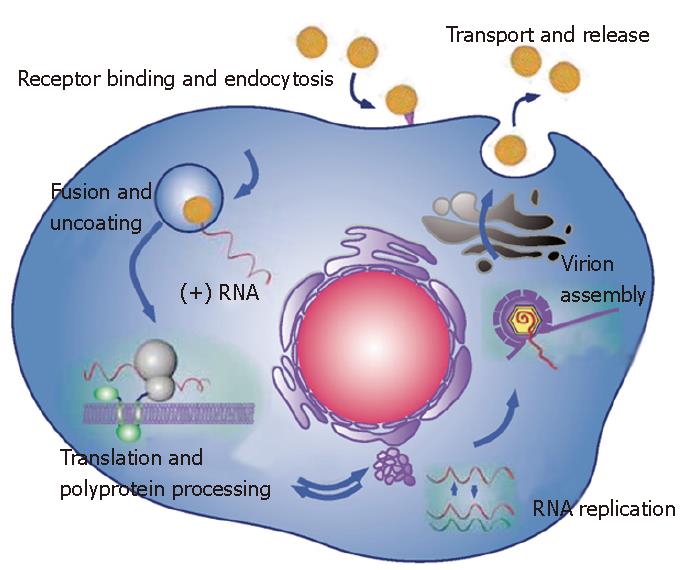
Figure 2 Schematic representation of the major steps of HCV life cycle.
The virus binds to a receptor at the cell surface, which leads to endocytosis of the particle. Fusion between the viral envelope and an endosomal membrane leads to the release of HCV genome into the cytosol. HCV genome is a positive strand RNA, which is directly translated and all the viral proteins are simultaneously produced. Expression of HCV proteins induces intracellular membrane alterations (the membranous web), which is the site of RNA replication. The nonstructural proteins NS3 to NS5B assemble in association with cellular factors to form a replication complex, which is responsible for RNA replication. Accumulation of HCV genomic RNA and the structural proteins leads to the assembly of a nucleocapsid, which acquires its envelope within an intracellular compartment. The viral particle is then secreted by following the classical secretory spathway.
- Citation: Dubuisson J. Hepatitis C virus proteins. World J Gastroenterol 2007; 13(17): 2406-2415
- URL: https://www.wjgnet.com/1007-9327/full/v13/i17/2406.htm
- DOI: https://dx.doi.org/10.3748/wjg.v13.i17.2406