Copyright
©2006 Baishideng Publishing Group Co.
World J Gastroenterol. Dec 7, 2006; 12(45): 7285-7291
Published online Dec 7, 2006. doi: 10.3748/wjg.v12.i45.7285
Published online Dec 7, 2006. doi: 10.3748/wjg.v12.i45.7285
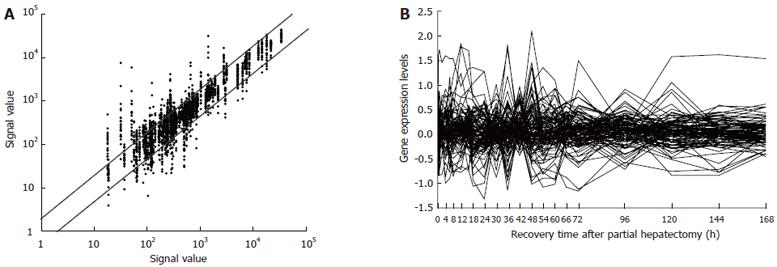
Figure 1 Expression frequency, abundance and changes of 107 genes associated with responses to chemical substances during rat liver regeneration.
Detection data of Rat Genome 230 2.0 array were analyzed and graphed by Microsoft Excel. A: Gene expression frequency. The dots above bias represent the genes up-regulated more than 2-fold, those under bias represent the ones down- more than 2-fold, and total times of up- and down-regulation were 441 and 221; the ones between two diagonal lines represent the genes without alteration in expression; B: Gene expression abundance and changes. Eighty-three genes were 2-128 folds up-regulated, and sixty genes 2-10 folds down-regulated.
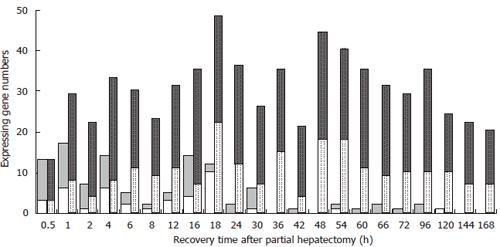
Figure 2 The initial and total expression profiles of 107 genes associated with responses to chemical substances at each time point of liver regeneration.
Grey bars: up-regulated genes; White bars: down-regulated genes. Blank bars represent initially expressing genes, in which up-regulation genes are predominantly in the forepart, prophase and early metaphase, and few genes initially expressed in the metaphase, early anaphase and middle anaphase, whereas no initial expression in the late anaphase. Dotted bars represent the total expressing genes, in which some genes are up-expressed, and the others down-expressed during LR.
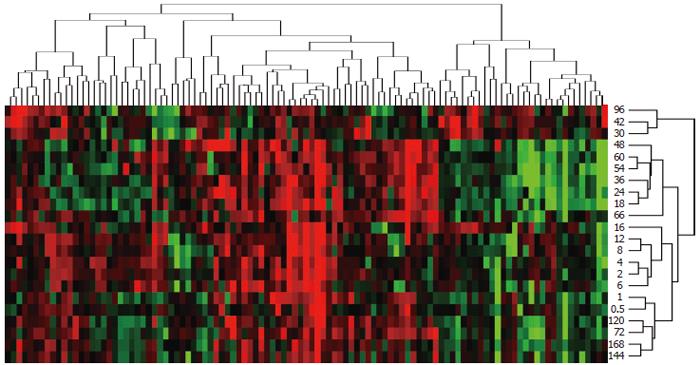
Figure 3 Expression similarity and time relevance clusters of 107 genes associated with responses to chemical substances during LR.
Detection data of Rat Genome 230 2.0 array were analyzed by H-clustering. Red represents up-regulated genes mainly associated with chemotaxis, xenobiotics metabolism, protein transport and apoptosis development; Green represents down- ones mainly associated with transportation and mucosal defense; Black: the genes having insignificant expression changes. The upper and right trees respectively show expression similarity and time series clusters, by which the above genes were classified into 5 and 14 groups.
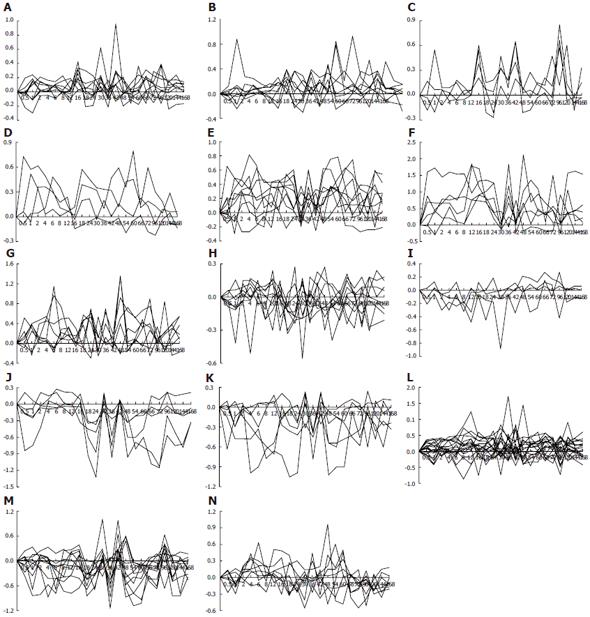
Figure 4 Gene expression patterns of 107 genes associated with responses to chemical substances during LR.
Twenty-six expression patterns were obtained by analysis of data detection of Rat Genome 230 2.0 array with Microsoft Excel. A-G: 47 up-regulated genes; H-K: 24 down-regulated genes; L-N: 36 up/down-regulated genes. X-axis represents recovery time after PH (h); Y-axis shows logarithm ratio of signal values of genes at each time point vs control.
- Citation: Qin SW, Zhao LF, Chen XG, Xu CS. Expression pattern and action analysis of genes associated with the responses to chemical stimuli during rat liver regeneration. World J Gastroenterol 2006; 12(45): 7285-7291
- URL: https://www.wjgnet.com/1007-9327/full/v12/i45/7285.htm
- DOI: https://dx.doi.org/10.3748/wjg.v12.i45.7285