Copyright
©2006 Baishideng Publishing Group Co.
World J Gastroenterol. Oct 7, 2006; 12(37): 6054-6058
Published online Oct 7, 2006. doi: 10.3748/wjg.v12.i37.6054
Published online Oct 7, 2006. doi: 10.3748/wjg.v12.i37.6054
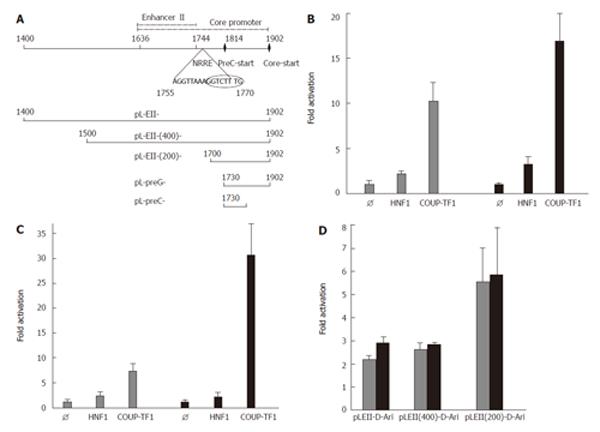
Figure 1 Structure (A) and activation of the precore (B), the pregenomic (C) and enhancer II (D) of HBV genotype D by COUP-TF1.
A: Schematic structure of HBV enhancer II and the core promoter. The sequence of the nuclear receptor response element (NRRE) and the natural deletion of nt 1763-1770 (encircled) is indicated. Below the transcriptional elements the HBV fragments cloned into the luciferase reporter vector pGL3 are shown. For cloning the indicated nucleotides from two plasmids containing wt (GenBank: Y07587) and HBV with a deletion of nt 1763-1770[18] were amplified and cloned. HepG2 cells were transfected with luciferase reporter constructs containing pregenomic promoter (pL-preG-D-Ari, wt in grey); B: Precore promoter (pL-preC-D-Ari, wt in grey) C: The complete and truncated wt enhancer II D: (pLEII-D-Ari, in grey) corresponding variant constructs with a deletion of nt 1763-1770 (Δ8bp) in black. For cotransfection the empty expression vector (Ø) or expression vectors for HNF1[27] or COUP-TF1[28] were employed.
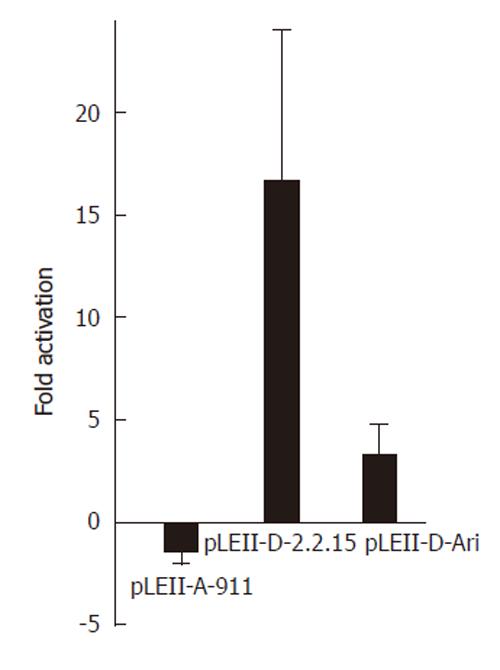
Figure 2 Influence of the nuclear receptor COUP-TF1 on enhancerII/core promoter constructs of different HBV genotypes.
HepG2 cells were transfected with luciferase constructs for the complete enhancer II (1400-1902) of genotype A (pLEII-A-991) or two constructs (pLEII-D-Ari and pLEII-D-2.2.15) derived from genotype D and the expression vector for COUP-TF1.
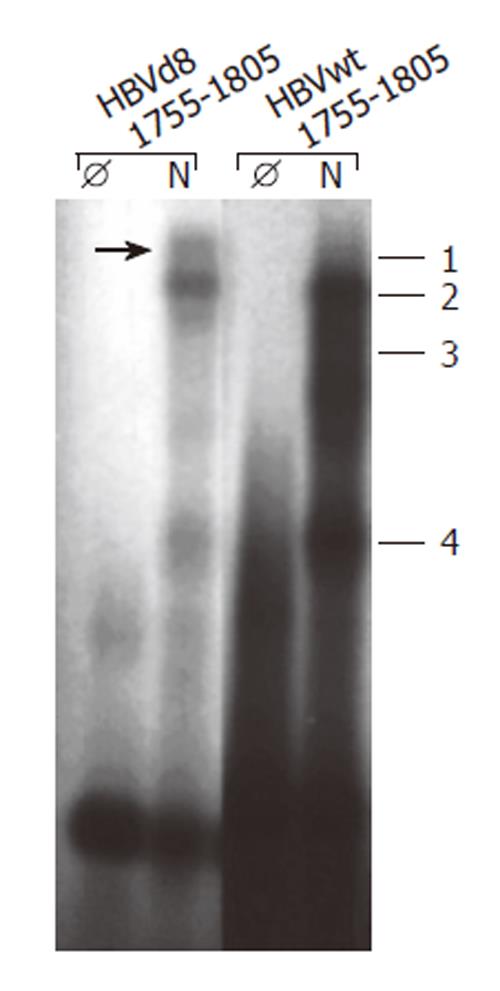
Figure 3 Deletion of nt 1763-1770 leads to altered band shifts in electrophoretic mobility shift assay with oligonucleotides nt 1755-1805.
EMSA was performed as described by[25] with oligonucleotide nt 1755-1805 wt (HBVwt 1755-1805) and with a deletion of nt 1763-1770 (HBVd8 1755-1805) incubated with (N) or without (Ø) nuclear extract prepared from HepG2 cells with a method[25].
- Citation: Fischer SF, Schmidt K, Fiedler N, Glebe D, Schüttler C, Sun J, Gerlich WH, Repp R, Schaefer S. Genotype-dependent activation or repression of HBV enhancer II by transcription factor COUP-TF1. World J Gastroenterol 2006; 12(37): 6054-6058
- URL: https://www.wjgnet.com/1007-9327/full/v12/i37/6054.htm
- DOI: https://dx.doi.org/10.3748/wjg.v12.i37.6054