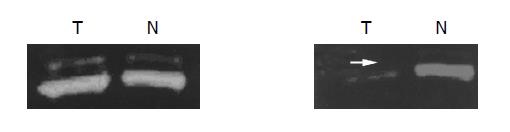Copyright
©2005 Baishideng Publishing Group Inc.
World J Gastroenterol. Mar 7, 2005; 11(9): 1333-1338
Published online Mar 7, 2005. doi: 10.3748/wjg.v11.i9.1333
Published online Mar 7, 2005. doi: 10.3748/wjg.v11.i9.1333
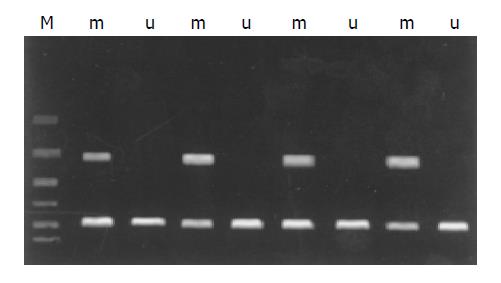
Figure 1 Methylation specific PCR (MSP) analysis of RASSF1A in extrahepatic cholangiocarcinoma with methylated DNA-specific primers (m) and unmethylated DNA-specific primers (u).
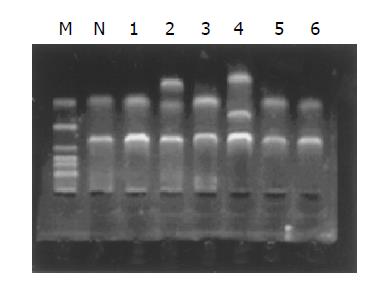
Figure 2 Restriction enzyme digestion of the promoter CpG island of bisulfite-treated in 4 tumors using Restriction enzyme TaqI.
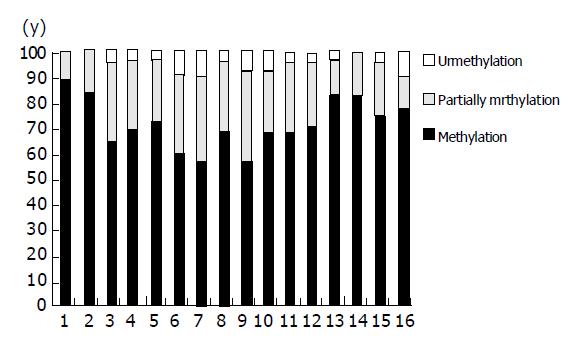
Figure 3 Frequency of methylation each CpG in RASSF1A promote in extrahepatic cholangiocarcinoma.
Figure 4 RASSF1A mutation analysis using SSCP in normal bile duct and 6 extrahepatic cholangiocarcinoma samples.
It displayed that tumor 2 has a extra band, tumor 3 has a extra band with shifting. We consider that there are mutation in those two tumor samples.
Figure 5 LOH analysis of microsatellite loci D3S1568 and D3S4604 at the region of chromosome 3p21.
3 using the primers labeled with FAM and HEX.
- Citation: Chen YJ, Tang QB, Zou SQ. Inactivation of RASSF1A, the tumor suppressor gene at 3p21.3 in extrahepatic cholangiocarcinoma. World J Gastroenterol 2005; 11(9): 1333-1338
- URL: https://www.wjgnet.com/1007-9327/full/v11/i9/1333.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i9.1333