Copyright
©2005 Baishideng Publishing Group Inc.
World J Gastroenterol. Mar 21, 2005; 11(11): 1634-1638
Published online Mar 21, 2005. doi: 10.3748/wjg.v11.i11.1634
Published online Mar 21, 2005. doi: 10.3748/wjg.v11.i11.1634
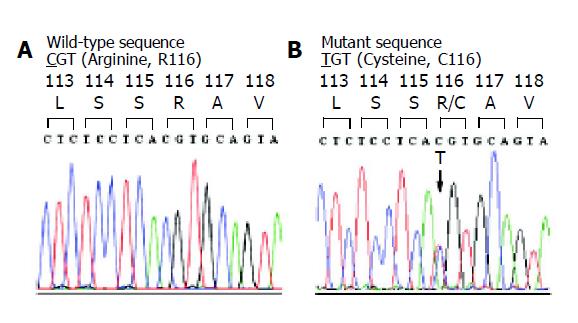
Figure 1 Chromatogram of DNA sequences in exon 3 of PRSS1 gene in a normal control (A) and a patient with chronic pancreatitis (B).
The tracing of the patient reveals a heterozygous missense mutation, g.2441C>T or c.346 C>T, which would result in a substitution of arginine (CGT) by cysteine (TGT) at amino acid residue 116 (R116C).
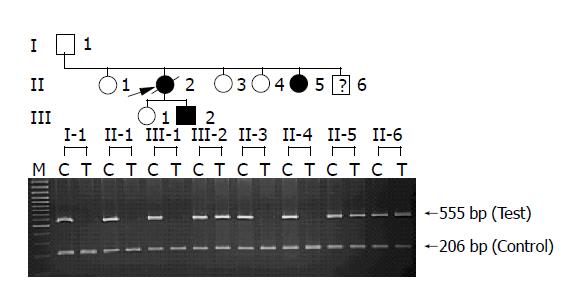
Figure 2 Allele specific amplification (ASA) analysis of R116C in the HP family.
In two affected (II-5 and III-2) and one unaffected (II-6) individuals, two bands (555 bp) amplified from both wild-type (C) and mutant (T) alleles are shown as heterozygous genotype, whereas only wild-type alleles (C) are present in the rest of unaffected individuals (I-1, II-1, II-3, II-4 and III-1). PCR products of 206 bp are shown as internal control in all lanes.
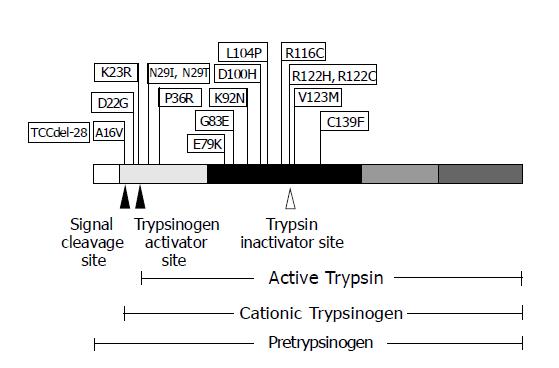
Figure 3 The schematic diagram of PRSS1 mutations discovered, to date, and their positions on the encoded protein.
PRSS1 protein is shown with five regions, each of which encoded by a different exon, depicted with varying shaded blocks. The arrowheads represent the proteolysis cleavage sites of the protein resulting in various forms of trypsin biogenesis.
-
Citation: Pho-Iam T, Thongnoppakhun W, Yenchitsomanus PT, Limwongse C. A Thai family with hereditary pancreatitis and increased cancer risk due to a mutation in
PRSS1 gene. World J Gastroenterol 2005; 11(11): 1634-1638 - URL: https://www.wjgnet.com/1007-9327/full/v11/i11/1634.htm
- DOI: https://dx.doi.org/10.3748/wjg.v11.i11.1634