Copyright
©The Author(s) 2004.
World J Gastroenterol. Feb 15, 2004; 10(4): 514-520
Published online Feb 15, 2004. doi: 10.3748/wjg.v10.i4.514
Published online Feb 15, 2004. doi: 10.3748/wjg.v10.i4.514
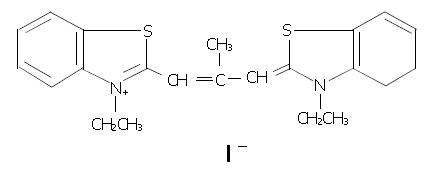
Figure 1 Chemical structure of 3, 3’-diethyl-9-methylthia-carbocyanine iodide (DMTCCI).
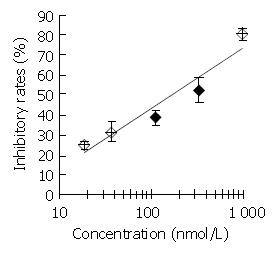
Figure 2 Inhibitory effect of DMTCCI on DNA primase activity.
Appropriate amounts of DMTCCI were incubated with the standard reaction mixture containing DNA primase-poly-merase complex purified from mice Ehrilich’s ascites carci-noma cells for 30 min at 37 °C. DNA primase reaction was mea-sured by the formation of RNA oligomers. Results were ex-pressed as mean ± SD for three independent experiments.
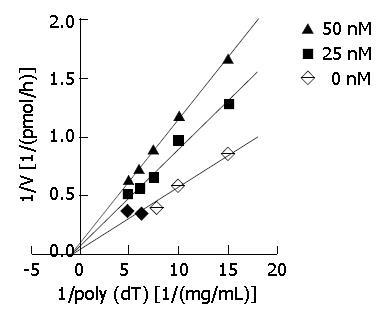
Figure 3 Kinetic analysis.
Primase activity was measured in the absence or presence of DMTCCI using the indicated concentrations of DNA template poly[dT]. A Dixon plot was made on the basis of the data. Basically similar results were obtained in three separate repetitions of the experiment.
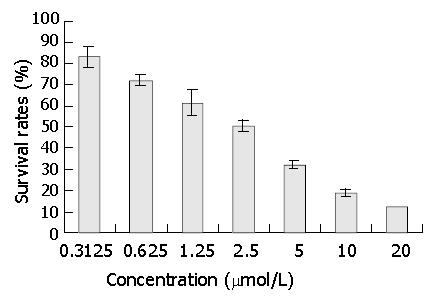
Figure 4 Effect of DNA primase inhibitor DMTCCI on proliferation of BEL-7402 cells.
BEL-7402 cells were treated with various concentrations of DMTCCI as indicated for 72 h. Cell viability was determined with MTT assay and shown as survival rate.
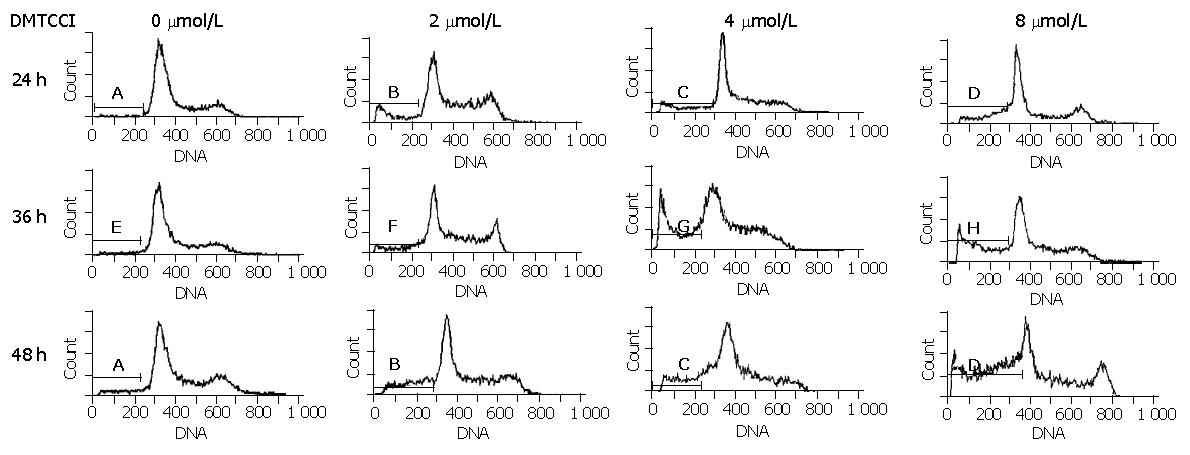
Figure 5 Flow cytometric analysis of BEL-7402 cells treated with DMTCCI.
Untreated cells (control) or cells treated with DMTCCI were stained with propidium iodide, their DNA content was measured by flow cytometry. The data shown are representative of three experiments performed.
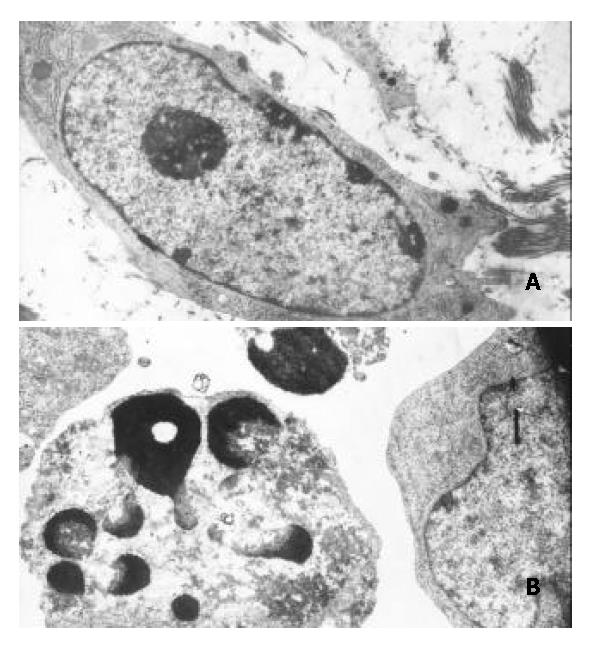
Figure 6 Morphological changes typical of apoptosis in BEL-7402 cells treated with 8 μmol/L of DMTCCI for 24 h.
BEL-7402 cells showed apoptotic morphological changes of the chro-matin being condensed and marginated. A: Control. × 5000; B: Treated with DMTTCI 8 μmol/L. × 7000.
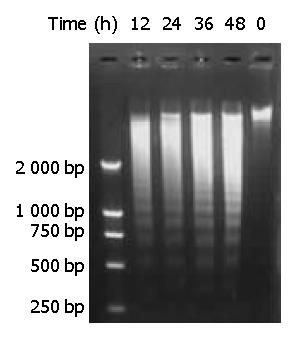
Figure 7 DNA fragmentation of BEL-7402 cells induced by DMTCCI.
BEL-7402 cells were incubated in the absence or pres-ence of DMTCCI 8 μM for 12 h, 24 h, 36 h, 48 h. Cells were harvested. DNA was isolated and DNA fragmentation was visualized as oligonucleosome-sized fragmentation in ethidium bromide after DNA agarose gel electrophoresis. This figure is a representative of three experiments.
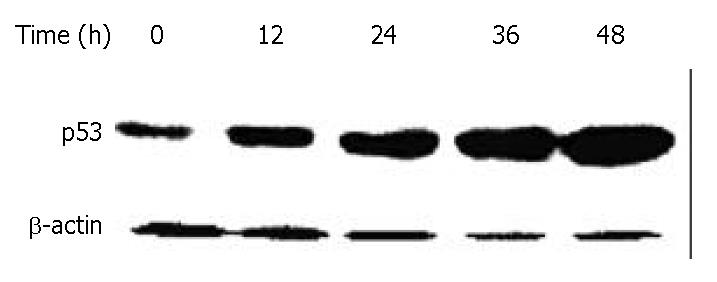
Figure 8 Down-regulation of p53 in BEL-7402 cells by DMTCCI.
BEL-7402 cells were incubated with 8 μM DMTCCI for indicated time. Cells were harvested, lysed and resolved in 10% SDS-polyacrylamide gel. p53 protein levels were determined by immunoblot analysis. β-actin was used as a control.
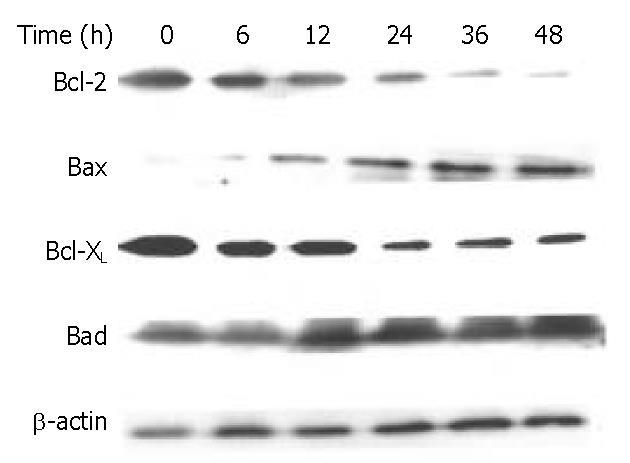
Figure 9 Effects of DMTCCI on levels of Bcl-2, Bax, Bcl-xL and Bad in BEL-7402 cells.
BEL-7402 cells were treated with DMTCCI (8 μM) for indicated time, total protein was isolated by immunoblotting analysis. After electrophoresis on a 10% SDS-polyacrylamide gel, the protein was transferred to a PVDF membrane. The membrane was analyzed by immunoblotting with anti-Bcl-2, Bax, Bcl-xL and Bad antibodies, respectively. β-actin protein was used as a control.
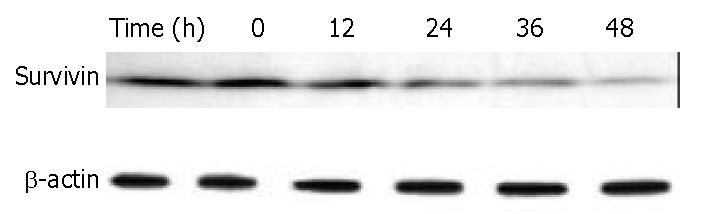
Figure 10 Down-regulation of survivin in BEL-7402 cells by DMTCCI.
BEL-7402 cells were incubated with 8 μM DMTCCI for indicated time. Cells were harvested, lysed and resolved in 10% SDS-polyacrylamide gel. Survivin protein levels were determined by immunoblot analysis. β-actin was used as a control.
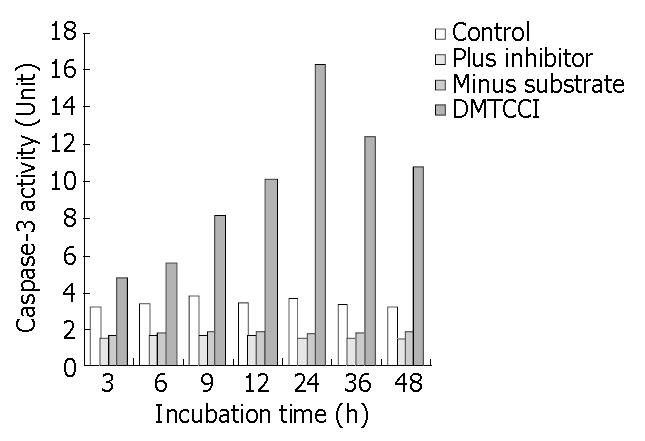
Figure 11 Inhibition of DMTCCI on activity of Caspase-3 in BEL-7402 cells.
BEL-7402 cells were harvested after exposure to 8 μmol/L of DMTCCI for indicated time. Activity of Caspase-3 was measured by ApoAlert caspase-3 assay kits. Cell lysates were prepared for each sample and used in the Caspase-3 ac-tivity assay using the colorimetric substrate DEVD-pNA. Ab-sorbance was read at 410 nm and This figure is a representa-tive of three experiments.
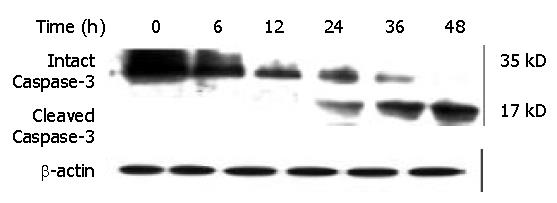
Figure 12 Effect of DMTCCI on levels of proteins of Caspase-3 in BEL-7402 cells.
BEL-7402 cells were treated with 8 μM DMTCCI for the indicated time. Western blot analysis was performed with anti-Caspase-3 antibody. β-actin was used as a control.
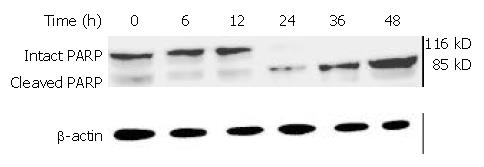
Figure 13 PARP cleavage in BEL-7402 cells induced by DMTCCI.
After BEL-7402 cells were treated with 8 μM DMTCCI for the indicated time, total protein was isolated by immunoblotting analysis. After electrophoresis on a 10% SDS-polyacrylamide gel, the protein was transferred to a PVDF membrane . The membrane was analyzed by immunoblotting with antibody against PARP. β-actin protein was used as a control.
- Citation: Li ZM, Liu ZC, Guan ZZ, Zhu XF, Zhou JM, Xie BF, Feng GK, Zhu ZY, Jiang WQ. Inhibition of DNA primase and induction of apoptosis by 3,3’-diethyl-9-methylthia-carbocyanine iodide in hepatocellular carcinoma BEL-7402 cells. World J Gastroenterol 2004; 10(4): 514-520
- URL: https://www.wjgnet.com/1007-9327/full/v10/i4/514.htm
- DOI: https://dx.doi.org/10.3748/wjg.v10.i4.514