Copyright
©The Author(s) 2004.
World J Gastroenterol. Oct 15, 2004; 10(20): 2967-2971
Published online Oct 15, 2004. doi: 10.3748/wjg.v10.i20.2967
Published online Oct 15, 2004. doi: 10.3748/wjg.v10.i20.2967
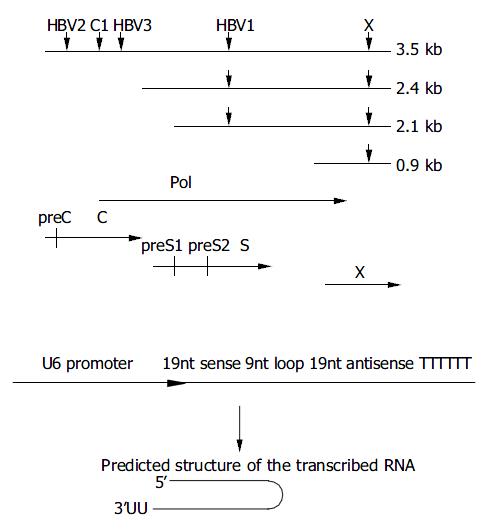
Figure 1 Design of siRNAs specific for HBV genome.
A: Down-ward arrows show the target sites within the RNA transcripts. The HBV ORFs are shown below aligned with HBV mRNA. B: Schematic presentation of U6 promoter constructs. The in-verted 19 nt sequences are separated by a 9 nt loop, transcrip-tion ends with 5Ts. The resulting shRNA (short hairpin RNA) are predicted to fold back to form a hairpin RNA.
Figure 2 Effect of RNAi on HBV protein expression in tran-siently transfected Huh-7 cells.
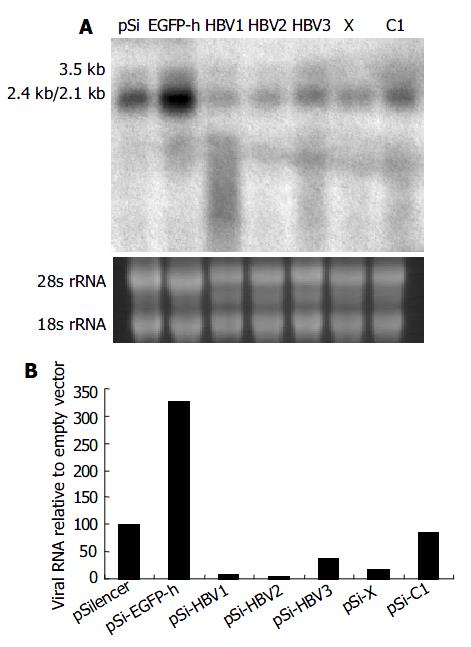
Figure 3 Effect of RNAi on HBV gene transcription detected by Northern blot.
A: Northern blot experiment of HBV transcripts. The 28 s and 18s rRNAs were visualized under ultraviolet light for equal loading control. B: The hybridiza-tion signal was quantitatively evaluated by SmartviewTM im-age analysis software (Shanghai Fu-ri Inc).
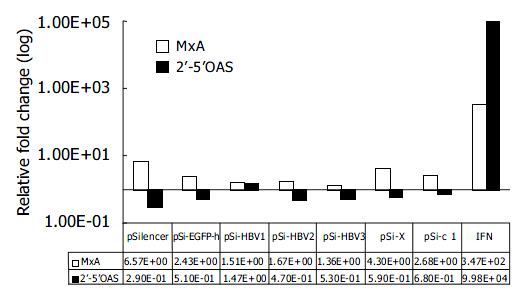
Figure 4 Effect of RNAi on IFN-induced genes.
Real-time PCR quantification of MxA and 2’-5’ OAS. The copy number of MxA and 2’-5’OAS were normalized with GAPDH. The rela-tive fold induction of each gene compared with mock trans-fection is shown.
- Citation: Zhang XN, Xiong W, Wang JD, Hu YW, Xiang L, Yuan ZH. siRNA-mediated inhibition of HBV replication and expression. World J Gastroenterol 2004; 10(20): 2967-2971
- URL: https://www.wjgnet.com/1007-9327/full/v10/i20/2967.htm
- DOI: https://dx.doi.org/10.3748/wjg.v10.i20.2967