Copyright
©The Author(s) 2019.
World J Clin Cases. Oct 6, 2019; 7(19): 2916-2929
Published online Oct 6, 2019. doi: 10.12998/wjcc.v7.i19.2916
Published online Oct 6, 2019. doi: 10.12998/wjcc.v7.i19.2916
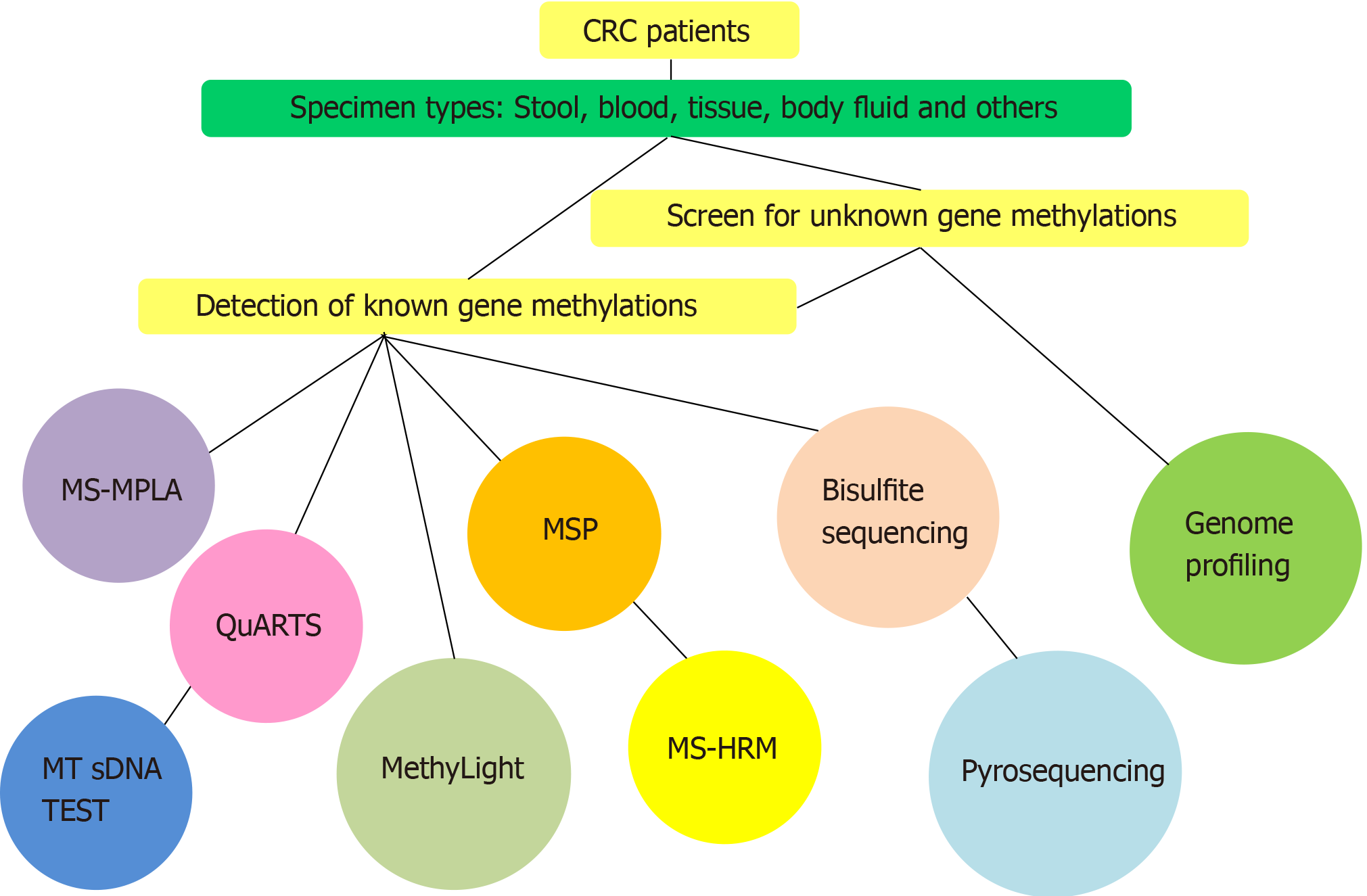
Figure 2 Diagram of the basic choices for methylation detection methods to study gene methylation in colorectal cancer.
In this diagram, we show the currently available methods and their different requirements for colorectal cancer assays. Researchers typically use two or more methods to investigate gene methylation. CRC: Colorectal cancer; MS-MLPA: Methylation-specific multiple ligation-dependent probe amplification; MS-HRM: Methylation-specific high resolution melting curve analysis; MSP: Methylation-specific polymerase chain reaction; MT-sDNA: Multitarget stool DNA test; QuARTS: Quantitative allele-specific real-time target and signal amplification assays.
- Citation: Zhan YX, Luo GH. DNA methylation detection methods used in colorectal cancer. World J Clin Cases 2019; 7(19): 2916-2929
- URL: https://www.wjgnet.com/2307-8960/full/v7/i19/2916.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v7.i19.2916