Published online Aug 16, 2018. doi: 10.12998/wjcc.v6.i8.224
Peer-review started: March 21, 2018
First decision: April 18, 2018
Revised: April 23, 2018
Accepted: May 11, 2018
Article in press: May 13, 2018
Published online: August 16, 2018
Peutz-Jeghers syndrome (PJS) is an autosomal dominant inherited disease, which is characterized by mucocutaneous pigmentation and multiple gastrointestinal hamartoma polyps. The germline mutation of LKB1/STK11 gene on chromosome 19p13.3 is considered to be the hereditary cause of PJS. However, must a patient with PJS have the LKB1/STK11 gene mutation? We here report a case of a male patient who had typical manifestations of PJS and a definite family history, but did not have LKB1/STK11 gene mutation. By means of high-throughput sequencing technology, only mutations in APC gene (c.6662T > C: p.Met2221Thr) and MSH6 gene (c.3488A > T: p.Glu1163Val) were detected. The missense mutations in APC and MSH6 gene may lead to abnormalities in structure and function of their expression products, and may result in the occurrence of PJS. This study suggests that some other genetic disorders may cause PJS besides LKB1/STK11 gene mutation.
Core tip: The germline mutation of LKB1/STK11 gene on chromosome 19p13.3 is considered to be the hereditary cause of Peutz-Jeghers syndrome (PJS). We report a male PJS patient, who had typical manifestations and a definite family history, and but did not have LKB1/STK11 gene mutation. By means of high-throughput sequencing technology, only mutations in APC gene (c.6662T > C: p.Met2221Thr) and MSH6 gene (c.3488A > T: p.Glu1163Val) were detected in this case. This study suggests that some other genetic disorders may cause PJS besides LKB1/STK11 gene mutation.
- Citation: Duan FX, Gu GL, Yang HR, Yu PF, Zhang Z. Must Peutz-Jeghers syndrome patients have the LKB1/STK11 gene mutation? A case report and review of the literature. World J Clin Cases 2018; 6(8): 224-232
- URL: https://www.wjgnet.com/2307-8960/full/v6/i8/224.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v6.i8.224
Peutz-Jeghers syndrome (PJS) is an autosomal dominant inherited disease characterized by mucocutaneous pigmentation and multiple gastrointestinal hamartoma polyps[1]. PJS is a very rare disease, with an incidence of about 1/25000[2]. It has prominent clinical manifestations, and serious clinical hazards since the gastrointestinal hamartoma polyps of PJS can lead to severe complications, such as gastrointestinal bleeding, bowel obstruction, intussusception and cancerization. PJS hamartoma polyps are mainly distributed in the small intestine (especially the proximal jejunum), so PJS has a persistent disease course and concealing lesion site[3-5]. This may cause mental stress and economic burden on the patients and their families. Therefore, it is necessary for us to study PJS.
The germline mutation of LKB1/STK11 gene on chromosome 19p13.3 is considered as the hereditary cause of PJS[6,7]. As a tumor suppressor gene, LKB1/STK11 contains 9 exons and 11 introns. At present, the detected mutation rate of LKB1/STK11 gene is 80%-94% by direct sequencing technology and multiplex ligation-dependent probe amplification (MLPA)[8-10]. This may suggest that the genetic etiology of PJS is heterogeneous[11-13]. Are there any other genetic causes for PJS besides LKB1/STK11 gene? Here, we report a case of male patient with PJS who had missense mutations of APC gene and MSH6 gene, but without mutations of LKB1/STK11 gene.
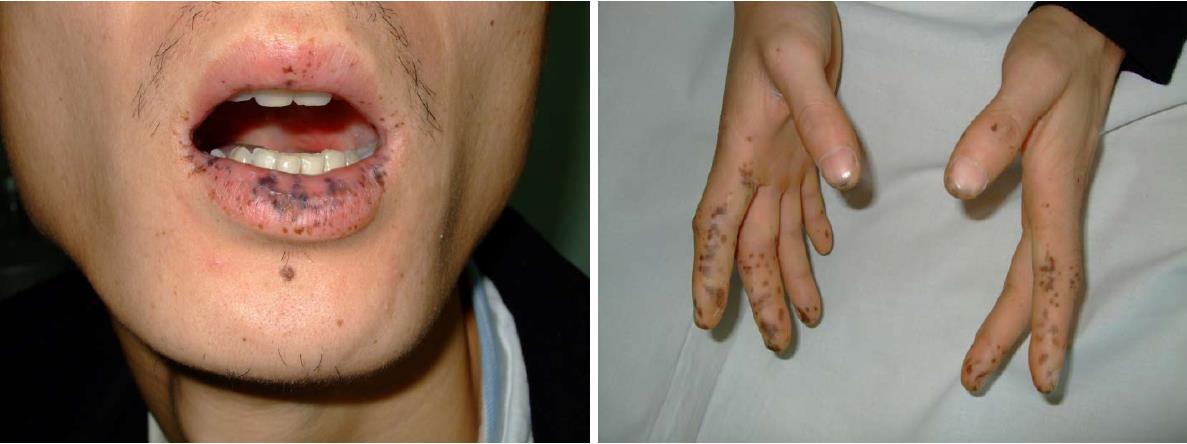
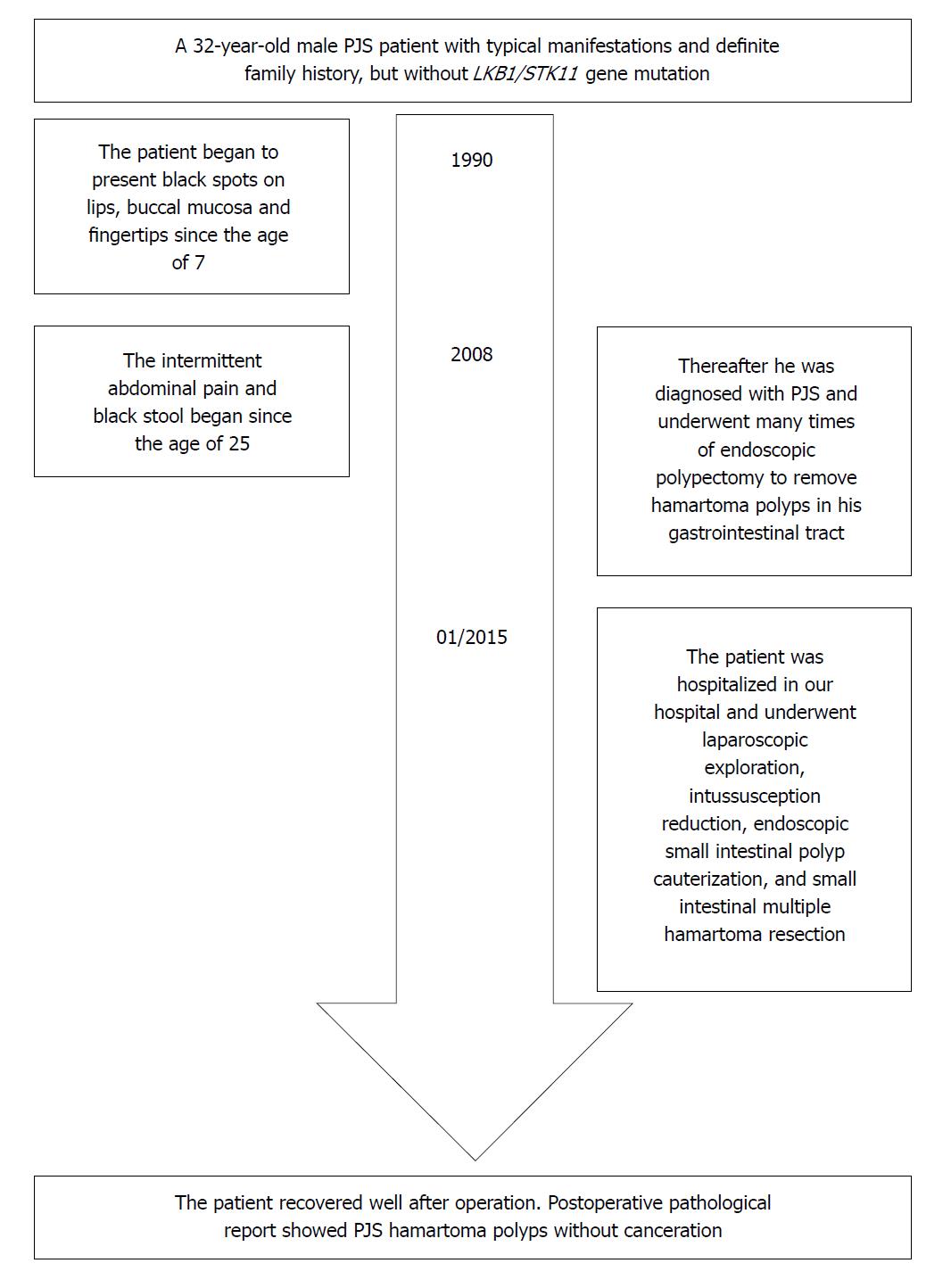
A 32-year-old male patient with PJS was hospitalized for endoscopic polypectomy in January 19, 2015. He had black spots on lips, buccal mucosa and fingertips since the age of 7 (Figure 1), and intermittent abdominal pain and black stool since the age of 25. He was diagnosed with PJS and underwent multiple endoscopic polypectomies to remove hamartoma polyps in the gastrointestinal tract via gastroscopy, enteroscopy and colonoscopy. His father was also diagnosed with PJS. This patient was in full compliance with the clinical diagnostic criteria for PJS issued by the WHO[14]: Patients without a family history can be diagnosed with PJS if they have three or more histopathologically confirmed PJS polyps, or characteristic, notable, mucocutaneous pigmentation with any amounts of PJS polyps. Patients with a family history can be diagnosed with PJS if they have any amounts of PJS polyps, or characteristic, notable, mucocutaneous pigmentation.
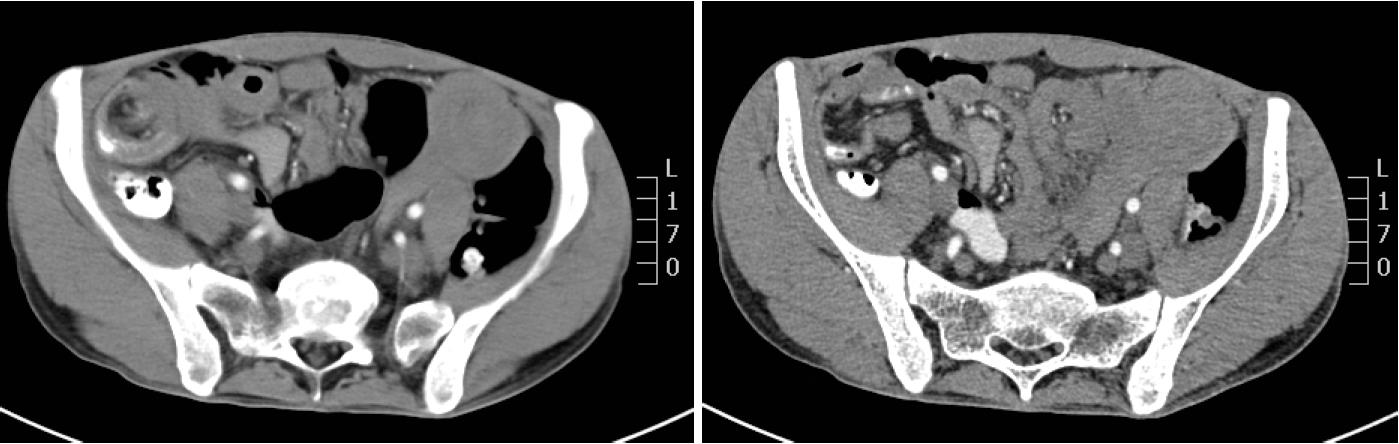
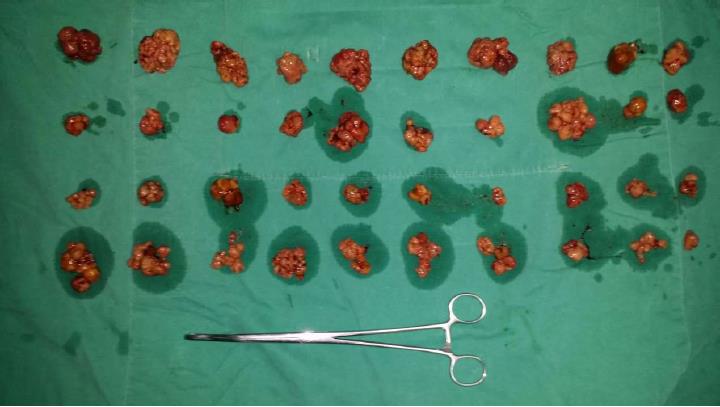
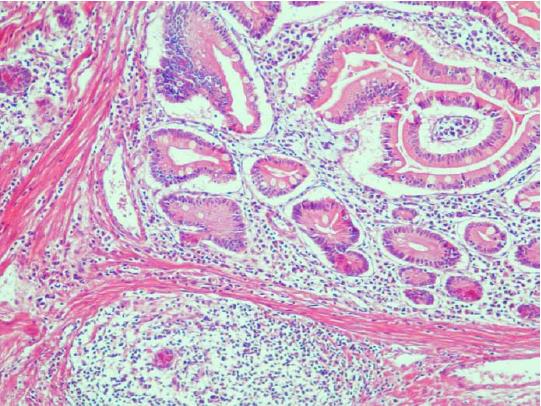
The abdominal and pelvic CT scan showed that there were many different sizes of polyps distributed in the descending duodenum and group 3rd-group 6th of the small intestine, and the local intestinal canal was torsional (Figure 2). Transanal double-balloon electronic enteroscopy revealed that there were multiple long pedicle, subpedicle or sessile polyps sized 0.5-1.2 cm in the colorectum, especially the descending colon, sigmoid colon and rectum. In addition, there were many huge lobulated polyps at a distance of 40 cm from the ileocecal valve that caused intestinal tract blockage. The patient underwent laparoscopic exploration, intussusception reduction, endoscopic small intestinal polyp cauterization, and small intestinal multiple hamartoma resection in January 25, 2015 (Figure 3). Postoperative pathological report showed PJS hamartoma polyps without canceration (Figures 4 and 5).
We used paraffin tissue DNA extraction kit to extract DNA from the polyp tissue according to the instructions of the kit (QIAamp Tissue DNA FFPE Tissue Kit, QIAGEN, QIAGEN Strasse 1407124 Hilden, QIAGEN). Preliminary screening of the sample DNA was carried out using the peak map, and purity test was done through the ultraviolet spectrophotometer (Nanodrop 2000, Thermo Scientific, Wilmington, DE, United States). Accurate quantitative analysis of complete DNA fragments in the sample DNA was performed using the fluorescence quantitative instrument (The Qubit 2 Fluorometer, Thermo Scientific, Wilmington, DE, United States). The results showed that the concentration of the sample DNA (A260/A280 2.01) was 1314 ng/μL, and the concentration of the complete DNA fragments was 160 ng/μL, which was higher than the requirement of 10 ng/μL.
We constructed cDNA libraries with Ion AmpliSeq Library Kit 2 according to the instructions of the manufacturer (Life 5791, Van Allen, Way Carlsbad. CA, United States). Ion Ampliseq Custom (IAD72340_182_pool 1, IAD72340_182_pool 2) (Life Technologies) was used to prepare a gene panel consisting of a primer pool for the entire coding region and exon-intron boundaries in 14 genes which often mutated in hereditary colorectal cancer (APC, AXIN2, BMPR1A, EPCAM, MLH1, MLH3, MSH2, MSH6, MUTYH, PMS1, PMS2, PTEN, SMAD4, LKB1/STK11). After amplification, the libraries were purified using paramagnetic particle method (AMPure XP reagent, Beckman, United States). The fluorescence quantitative PCR instrument (ViiA 7 Dx, Life, Technologies Holdings Pte Ltd Block, Singapore) was used to detect the quantitative index of the libraries. Then the template was dealt with Ion OneTouch 2 template system (Life Technologies, Carlsbad, CA, United States) for preparation (Ion OneTouch2) and enrichment (Ion OneTouch ES). Finally, the high-throughput sequencing was performed on Ion PGM (Life, Technologies Holdings Pte Ltd Block, Singapore) using Ion 316 CHIP (Life Technologies Corp, Carlsbad, CA, United States).
The quality control parameters of the sequencing data were as follows: percentage reads on target > 85%, uniformity of base coverage > 85%, average base coverage depth: 200 ×-500 ×. The data were analyzed by Torrent Suite software (Life Technologies, v5.0.4) and compared with database of hg19 human reference genome. The detected gene mutations were annotated by Ion Reporter software (https://ionreporter.lifetechnologies.com/ir/secure/home.html) and ANNOVAR package (http://wannovar.wglab.org/).
Preliminary screening of mutation sites for verification was carried out according to the mutation frequency. Allele frequency database of the herd was based on the dbSNP database (http://www.ncbi.nlm.nih.gov/projects/SNP/), 1000 Genomes Project (http://ftp.ncbi.nih.gov/) and the genome aggregation database (gnomAD, http://gnomad.broadinstitute.org/). According to the HGMD (version 2017.03, http://www.hgmd.cf.ac.uk/ac/index.php), mutation sites of minor allele frequency < 0.01 and the suspected or pathogenetic sites with frequency of 0.01-0.05 were retained for verification.
The Sanger sequencing technology was applied to candidate sites verification. Protein function was predicted with the software Polymorphism Phenotyping V2 (PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/index.shtml), MutationTaster (http://www.mutationtaster.org/), functional analysis was performed using the Hidden Markov Models (FATHMM, http://fathmm.biocompute.org.uk/index.html) and Mendelian Clinically Applicable Pathogenicity (M-CAP, http://bejerano.stanford.edu/MCAP/). GERP++(http://mendel.stanford.edu/SidowLab/downloads/gerp/index.html) and PhyloP (http://compgen.bscb.cornell.edu/phast) were used to forecast the conservatism of the impaired amino-acid residues.
This sequencing could read 28269827 bases. The base number of Q20 was 24754457, accounting for 87.56%. Percentage reads on target was 92.12%. Average base coverage depth was 286 × (coverage ≥ 20 × 94.39%, coverage ≥ 100 × 77.38%, coverage ≥ 500 × 12.75%). Uniformity of base coverage was 92.12%.
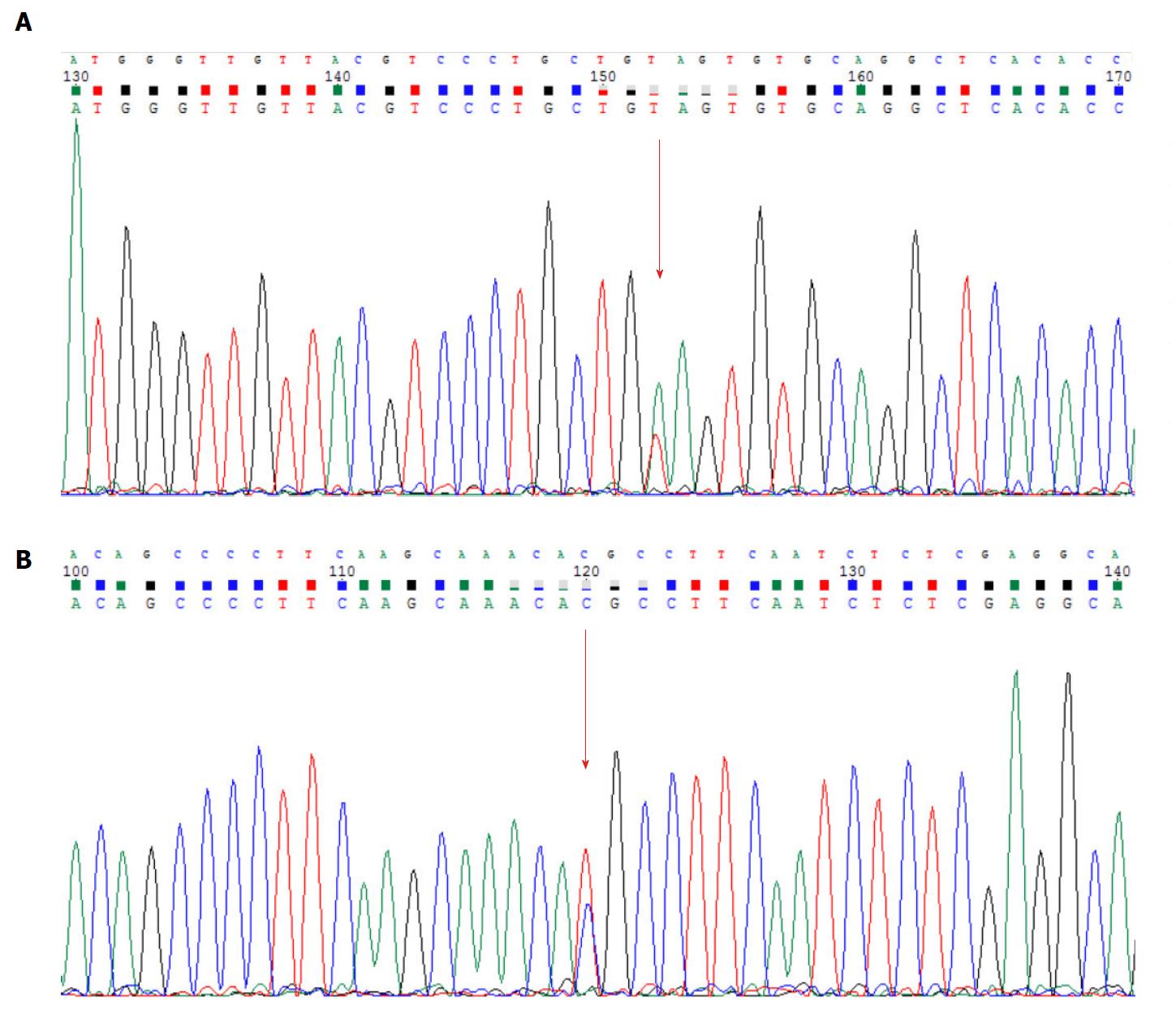
No mutation of LKB1/STK11 gene was found in this patient, but he carries missense mutations in exon 6 of MSH6 gene and exon 16 of APC gene (MSH6: NM_000179.2: exon6: c.3488A>T: p.Glu1163Val; APC: NM_000038.5: exon16: c.6662T>C: p.Met2221Thr) (Figure 6). The Sanger sequencing results proved the existence of these mutations (primers: APC: forward primer: CCAGGGGAGAAAAGTACATTGGA, reverse primer: ACTTGTACTTGAGGAGCTATTTCG; MSH6: forward primer: ACAGAACCAACGTACATGTGA, reverse primer: TTCTGTCTGAGGCACCAAGTC).
The changes in the structure and function of APC protein caused by gene mutations were analyzed and determined as benign by Polyphen-2, as disease caused by MutationTaster, and as damage by FATHMM and by M-CAP. Evolutionary conservative analysis software of GERP++ and phyloP showed that the impaired amino-acid residue was conserved in different species, suggesting that the mutation may be pathogenic.
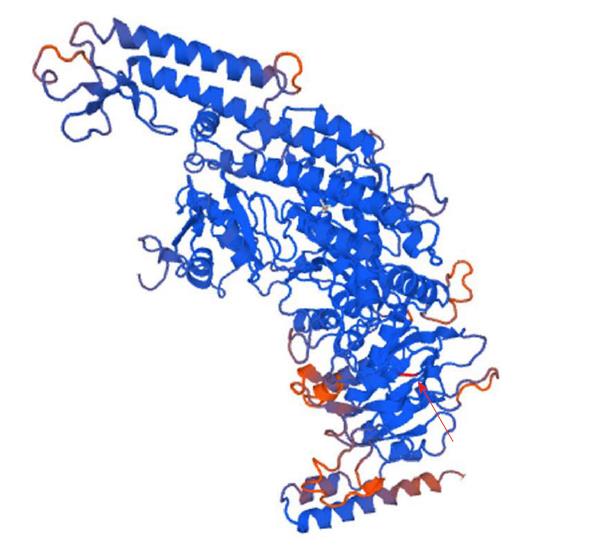
The structure and function changes of the mutated MSH6 protein were analyzed and defined as possible damage or benign by Polyphen-2, as disease caused by MutationTaster and as damage by FATHMM. Evolutionary conservative analysis software of GERP++ and phyloP indicated that the mutant amino-acid residue was most evolutionarily conserved and was likely pathogenic (Tables 1 and 2).
| Gene | Polyphen-2_HDIV | Polyphen-2_HVAR | MutationTaster | FATHMM | M-CAP | |||||
| Score | Prediction | Score | Prediction | Score | Prediction | Score | Prediction | Score | Prediction | |
| MSH6 | 0.670 | Possibly damaging | 0.411 | Benign | 1 | Disease causing | -2.12 | Damaging | - | - |
| APC | 0.156 | Benign | 0.026 | Benign | 0.737 | Disease causing | -2.47 | Damaging | 0.046 | Damaging |
| Gene | Exon | Protein | Coding | GERP++ | PhyloP | ||
| Score | Prediction | Score | Prediction | ||||
| MSH6 | 6 | p.Glu1163Val | c.3488A > T | 5.23 | Conserved | 8.923 | Conserved |
| APC | 16 | p.Met2221Thr | c.6662T > C | 6.02 | Conserved | 3.925 | Conserved |
In this study, the patient had a clear family history and typical clinical manifestations of PJS, and histologic examination of his polyps proved to be hamartoma. However, we only detected the missense mutations in the APC gene and MSH6 gene from his polyp tissue by high-throughput sequencing and Sanger sequencing technology, but no mutation in the LKB1/STK11 gene. The mutation detected in APC gene has not been reported previously, and the MAF was 8.3e-6 according to the ExAC database. The mutation detected in MSH6 gene has been reported in some sequencings[15-17], the MAF was 0.0012 according to the ExAC database as well as 0.0028 from 1000G database.
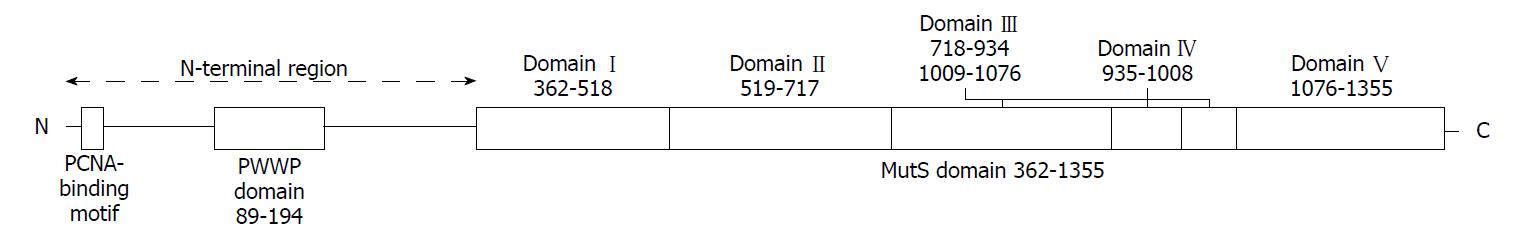
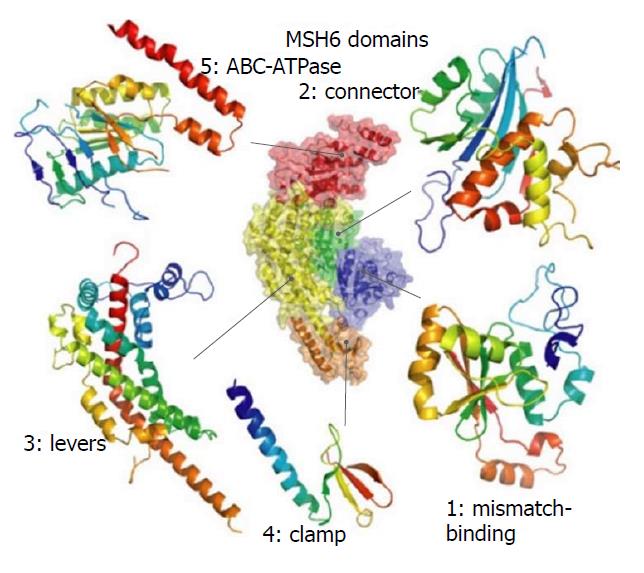
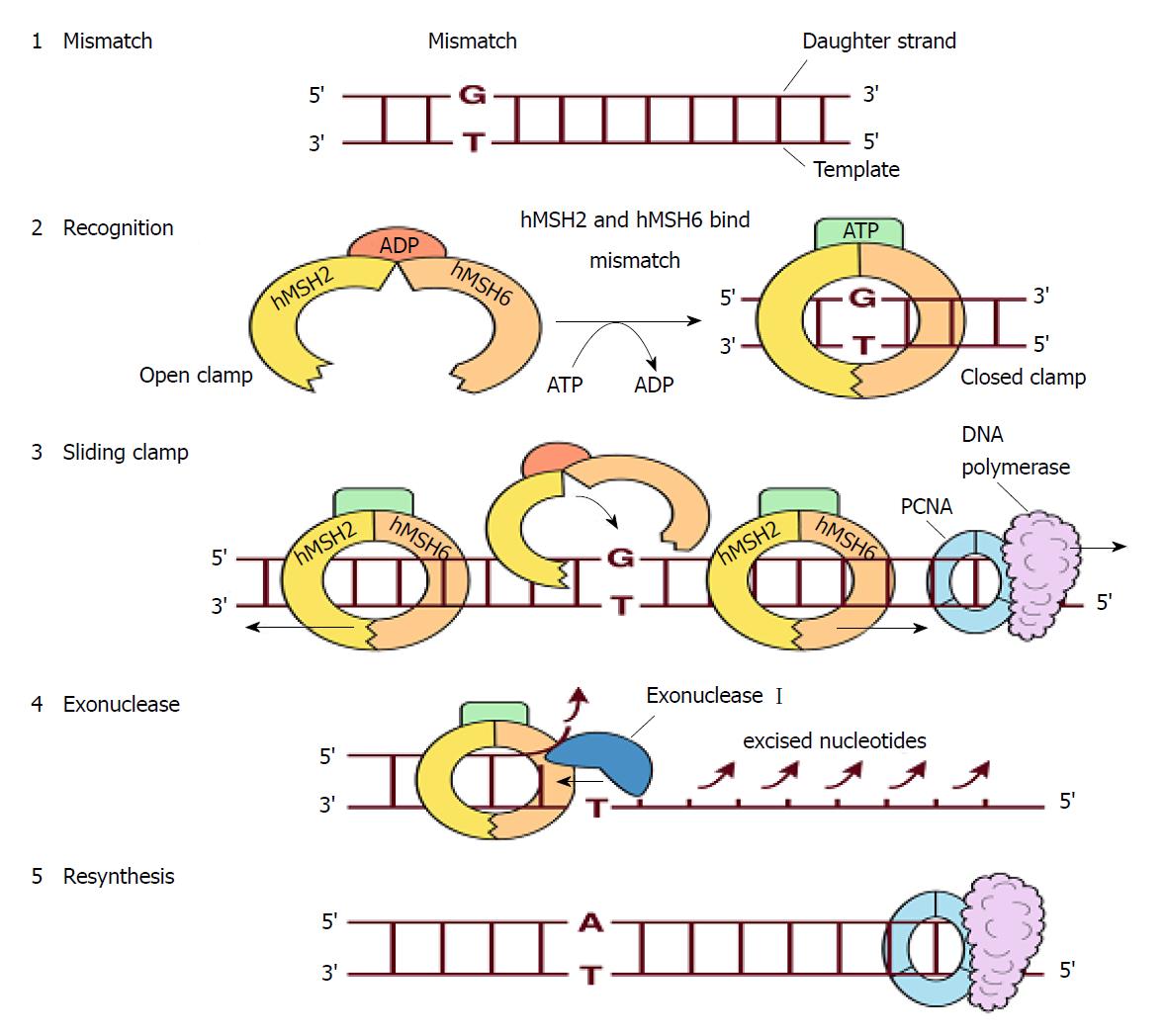
The DNA mismatch repair proteins (MMR), including MLH1, MSH2, MSH3, MSH6, PMS1 and PMS2, could remain gene stability by means of discovering and repairing mismatched bases and insertion/deletion loops improperly incorporated during DNA synthesis[18-21]. Expression deletion of MMR protein can be found in about 15%-25% sporadic tumors of various tissues[22]. The MSH6 gene is located on chromosome 2p16.3, which contains 24 000 base pairs and 10 exons. The MSH6 protein was first reported as G/T mismatch repair binding protein (GTBP) in 1995, it constitutes MutSα complex with MSH2[23-25]. The main domains of MSH6 protein from N-terminal to C-terminal included (Figure 7): PCNA-binding motif, PWWP domain and the MutS domain[26-28]. The MutS domain is composed of five parts: (1) Mismatch binding domain, including the 362-518th amino acids; (2) connector domain, including the 519-717th amino acids; (3) levers domain, including the 718-934th and 1009-1075th amino acids; (4) clamp domain, including the 935-1008th amino acids; (5) ABC-ATPase domain, including the 1076-1355th amino acids[29] (Figure 8). The gene mutation (c.3488A>T) detected in our patient led the 1163rd amino acid of glutamate to become valine in the MSH6 protein (Figure 9). Changing an acidic amino-acid into a non-polar hydrophobic amino-acid may cause the ABC-ATPase domain conformational change and affect the hydrolysis of ATP or ADP to ATP conversion process. This change could disorder the open and close conformational transition of the MutSα complex and eventually lead to the loss of MMR expression (Figure 10).
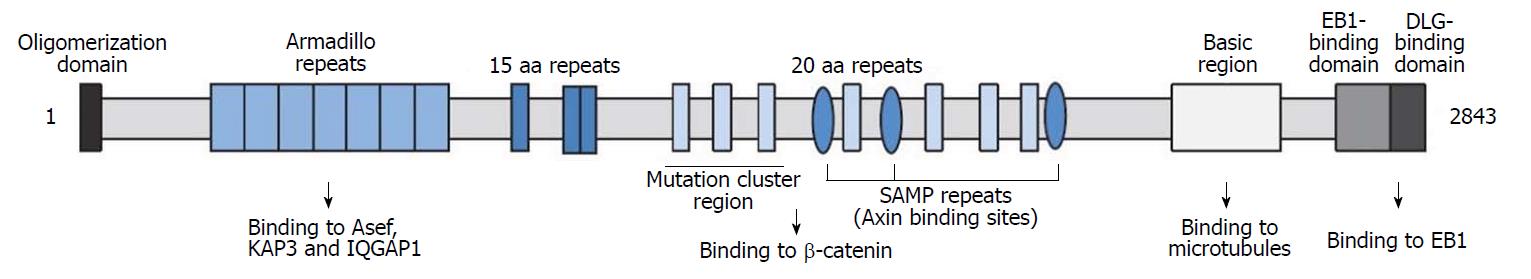
The APC gene is located on the autosome 5q21. Its main functions include inhibiting cell proliferation, promoting differentiation, promoting apoptosis, participating in cell migration, cell adhesion and chromosome separation. It is generally believed that the mutation of APC gene is an important initiator for the occurrence and development of colorectal cancer[30,31]. Mutated APC can also be seen in some brain and breast cancers[32,33]. The molecular weight of APC protein is about 312 kDa, and its N-terminal domains mainly include: (1) Oligomerization domain; (2) armadillo repeats domain; (3) 15-amino acid or 20-amino acid repeats domain; (4) SAMP repeats domain and Mutation cluster region (MCR); (5) Basic region for binding microtubule or f-actin, including the 2219-2580th amino acids; (6) EB1-binding domain; (7) DLG-binding domain[34,35] (Figure 11). APC protein can participate in the composition of a complex promoting the destruction of β-catenin (a transcriptional permissive factor), and then negatively regulate the classical Wnt signal pathway. Its C-terminal region, especially the 2200-2400th amino-acid, is rich in bases that could boost the binding process of microtubules[36-38]. APC protein of cell periphery plays an important role in binding microtubule network, establishing cell polarity and inhibiting invasion according to many studies[39,40]. A research based on a zebrafish model suggests that cell differentiation may be related to the Warburg effect mediated by APC[41]. In addition, the differentiation mechanism of human intestinal epithelial cells may be similar to that of zebrafish[42]. In this case, the mutation (c.6662T>C) causes 2221st methionine of the APC protein to become threonine, which may result in the protein conformation change, and affect cell differentiation, adhesion and migration.
In summary, we detected MSH6 and APC gene mutations in a case of PJS by high-throughput sequencing technology, but failed to detect mutations in LKB1/STK11 gene, and the mutation of APC gene (c.6662T>C: p.Met2221Thr) had never been reported previously. This suggests that mutations of MSH6 and APC genes may affect the occurrence and development of PJS by affecting the MMR pathway, cell differentiation, adhesion and migration. Our research may be helpful in expanding the mutation spectrum of PJS and revealing its genetic heterogeneity.
A 32-year-old male patient with Peutz-Jeghers syndrome (PJS).
Peutz-Jeghers syndrome.
Multiple long pedicle, subpedicle or sessile polyps sized 0.5-1.2 cm appeared in the colorectum, especially the descending colon, sigmoid colon and rectum.
Many polyps of different sizes were distributed in the descending duodenum and the 3rd to 6th segments of the small intestine, and the local intestinal canal was torsional.
The patient underwent a combined laparoscopic and endoscopic surgery.
PJS hamartoma polyps without canceration.
Instead of LKB1/STK11 gene, we detected missense mutations of APC gene and MSH6 gene.
This research may be helpful in expanding the mutation spectrum of PJS and revealing its genetic heterogeneity.
CARE Checklist (2013) statement: We have read the CARE Checklist (2013), and the manuscript was prepared and revised according to the CARE Checklist (2013).
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): A
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Luca F, Lee MW, Mulvihill SJ S- Editor: Wang XJ L- Editor: Ma JY E- Editor: Tan WW
| 1. | Beggs AD, Latchford AR, Vasen HF, Moslein G, Alonso A, Aretz S, Bertario L, Blanco I, Bülow S, Burn J. Peutz-Jeghers syndrome: a systematic review and recommendations for management. Gut. 2010;59:975-986. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 516] [Cited by in F6Publishing: 414] [Article Influence: 29.6] [Reference Citation Analysis (0)] |
| 2. | Lu XJ, Gu GL, Wei XM, Ren L, Ning SB, Li DC. Expression of key members of classical Wnt signal pathway in Peutz-Jeghers syndrome. World Chinese Journal of Digestology. 2013;21:655-660. [DOI] [Cited in This Article: ] [Cited by in CrossRef: 1] [Cited by in F6Publishing: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 3. | Wang H, Luo T, Liu WQ, Huang Y, Wu XT, Wang XJ. Clinical presentations and surgical approach of acute intussusception caused by Peutz-Jeghers syndrome in adults. J Gastrointest Surg. 2011;15:2218-2225. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 24] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 4. | Wang SL, Gu GL, Mao GP. The experience of diagnosis and treatment gastrointestinal polyps in 36 cases with Peutz-Jeghers syndrome. Zhonghua Weichang Waike Zazhi. 2009;12:428. [DOI] [Cited in This Article: ] |
| 5. | Latchford AR, Phillips RK. Gastrointestinal polyps and cancer in Peutz-Jeghers syndrome: clinical aspects. Fam Cancer. 2011;10:455-461. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in F6Publishing: 24] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 6. | Jenne DE, Reimann H, Nezu J, Friedel W, Loff S, Jeschke R, Müller O, Back W, Zimmer M. Peutz-Jeghers syndrome is caused by mutations in a novel serine threonine kinase. Nat Genet. 1998;18:38-43. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 802] [Cited by in F6Publishing: 845] [Article Influence: 32.5] [Reference Citation Analysis (0)] |
| 7. | Hemminki A, Markie D, Tomlinson I, Avizienyte E, Roth S, Loukola A, Bignell G, Warren W, Aminoff M, Höglund P. A serine/threonine kinase gene defective in Peutz-Jeghers syndrome. Nature. 1998;391:184-187. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1136] [Cited by in F6Publishing: 1058] [Article Influence: 40.7] [Reference Citation Analysis (0)] |
| 8. | Aretz S, Stienen D, Uhlhaas S, Loff S, Back W, Pagenstecher C, McLeod DR, Graham GE, Mangold E, Santer R. High proportion of large genomic STK11 deletions in Peutz-Jeghers syndrome. Hum Mutat. 2005;26:513-519. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 183] [Cited by in F6Publishing: 199] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 9. | Volikos E, Robinson J, Aittomäki K, Mecklin JP, Järvinen H, Westerman AM, de Rooji FW, Vogel T, Moeslein G, Launonen V. LKB1 exonic and whole gene deletions are a common cause of Peutz-Jeghers syndrome. J Med Genet. 2006;43:e18. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 104] [Cited by in F6Publishing: 112] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 10. | de Leng WW, Jansen M, Carvalho R, Polak M, Musler AR, Milne AN, Keller JJ, Menko FH, de Rooij FW, Iacobuzio-Donahue CA. Genetic defects underlying Peutz-Jeghers syndrome (PJS) and exclusion of the polarity-associated MARK/Par1 gene family as potential PJS candidates. Clin Genet. 2007;72:568-573. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36] [Cited by in F6Publishing: 37] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 11. | Mehenni H, Gehrig C, Nezu J, Oku A, Shimane M, Rossier C, Guex N, Blouin JL, Scott HS, Antonarakis SE. Loss of LKB1 kinase activity in Peutz-Jeghers syndrome, and evidence for allelic and locus heterogeneity. Am J Hum Genet. 1998;63:1641-1650. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 139] [Cited by in F6Publishing: 152] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 12. | Tomlinson IP, Olschwang S, Abelovitch D, Nakamura Y, Bodmer WF, Thomas G, Markie D. Testing candidate loci on chromosomes 1 and 6 for genetic linkage to Peutz-Jeghers’ disease. Ann Hum Genet. 1996;60:377-384. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 16] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 13. | Zhao XR, Kang LC, Zhou YS, Jia YX, Chen Z, Kang SH, Li WM, Zhao M, Cui JT, Sun AL. [Mutations of fragile histidine triad gene in Peutz-Jeghers syndrome and canceration]. Ai Zheng. 2003;22:50-54. [PubMed] [Cited in This Article: ] |
| 14. | Aaltonen LA. Hereditary intestinal cancer. Semin Cancer Biol. 2000;10:289-298. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in F6Publishing: 23] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 15. | Duzkale H, Shen J, McLaughlin H, Alfares A, Kelly MA, Pugh TJ, Funke BH, Rehm HL, Lebo MS. A systematic approach to assessing the clinical significance of genetic variants. Clin Genet. 2013;84:453-463. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 113] [Cited by in F6Publishing: 130] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 16. | Bodian DL, McCutcheon JN, Kothiyal P, Huddleston KC, Iyer RK, Vockley JG, Niederhuber JE. Germline variation in cancer-susceptibility genes in a healthy, ancestrally diverse cohort: implications for individual genome sequencing. PLoS One. 2014;9:e94554. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 67] [Cited by in F6Publishing: 67] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 17. | Chang YC, Chang JG, Liu TC, Lin CY, Yang SF, Ho CM, Chen WT, Chang YS. Mutation analysis of 13 driver genes of colorectal cancer-related pathways in Taiwanese patients. World J Gastroenterol. 2016;22:2314-2325. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 16] [Cited by in F6Publishing: 17] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 18. | Haugen AC, Goel A, Yamada K, Marra G, Nguyen TP, Nagasaka T, Kanazawa S, Koike J, Kikuchi Y, Zhong X. Genetic instability caused by loss of MutS homologue 3 in human colorectal cancer. Cancer Res. 2008;68:8465-8472. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 116] [Cited by in F6Publishing: 121] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 19. | Woerner SM, Tosti E, Yuan YP, Kloor M, Bork P, Edelmann W, Gebert J. Detection of coding microsatellite frameshift mutations in DNA mismatch repair-deficient mouse intestinal tumors. Mol Carcinog. 2015;54:1376-1386. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 29] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 20. | Mello JA, Acharya S, Fishel R, Essigmann JM. The mismatch-repair protein hMSH2 binds selectively to DNA adducts of the anticancer drug cisplatin. Chem Biol. 1996;3:579-589. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 133] [Cited by in F6Publishing: 143] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 21. | Negureanu L, Salsbury FR. Insights into protein - DNA interactions, stability and allosteric communications: a computational study of mutSα-DNA recognition complexes. J Biomol Struct Dyn. 2012;29:757-776. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 19] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 22. | Peltomäki P. Role of DNA mismatch repair defects in the pathogenesis of human cancer. J Clin Oncol. 2003;21:1174-1179. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 508] [Cited by in F6Publishing: 487] [Article Influence: 23.2] [Reference Citation Analysis (0)] |
| 23. | Drummond JT, Li GM, Longley MJ, Modrich P. Isolation of an hMSH2-p160 heterodimer that restores DNA mismatch repair to tumor cells. Science. 1995;268:1909-1912. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 427] [Cited by in F6Publishing: 460] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 24. | Papadopoulos N, Nicolaides NC, Liu B, Parsons R, Lengauer C, Palombo F, D’Arrigo A, Markowitz S, Willson JK, Kinzler KW. Mutations of GTBP in genetically unstable cells. Science. 1995;268:1915-1917. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 333] [Cited by in F6Publishing: 358] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 25. | Palombo F, Gallinari P, Iaccarino I, Lettieri T, Hughes M, D’Arrigo A, Truong O, Hsuan JJ, Jiricny J. GTBP, a 160-kilodalton protein essential for mismatch-binding activity in human cells. Science. 1995;268:1912-1914. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 378] [Cited by in F6Publishing: 403] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 26. | Terui H, Akagi K, Kawame H, Yura K. CoDP: predicting the impact of unclassified genetic variants in MSH6 by the combination of different properties of the protein. J Biomed Sci. 2013;20:25. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 11] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 27. | Gassman NR, Clodfelter JE, McCauley AK, Bonin K, Salsbury FR Jr, Scarpinato KD. Cooperative nuclear localization sequences lend a novel role to the N-terminal region of MSH6. PLoS One. 2011;6:e17907. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 19] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 28. | Zhang J, Wang X, de Voer RM, Hehir-Kwa JY, Kamping EJ, Weren RDA, Nelen M, Hoischen A, Ligtenberg MJL, Hoogerbrugge N. A molecular inversion probe-based next-generation sequencing panel to detect germline mutations in Chinese early-onset colorectal cancer patients. Oncotarget. 2017;8:24533-24547. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 12] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 29. | Warren JJ, Pohlhaus TJ, Changela A, Iyer RR, Modrich PL, Beese LS. Structure of the human MutSalpha DNA lesion recognition complex. Mol Cell. 2007;26:579-592. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 267] [Cited by in F6Publishing: 270] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 30. | Boland CR, Koi M, Chang DK, Carethers JM. The biochemical basis of microsatellite instability and abnormal immunohistochemistry and clinical behavior in Lynch syndrome: from bench to bedside. Fam Cancer. 2008;7:41-52. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 129] [Cited by in F6Publishing: 129] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 31. | Narayan S, Roy D. Role of APC and DNA mismatch repair genes in the development of colorectal cancers. Mol Cancer. 2003;2:41. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 118] [Cited by in F6Publishing: 129] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 32. | Hamilton SR, Liu B, Parsons RE, Papadopoulos N, Jen J, Powell SM, Krush AJ, Berk T, Cohen Z, Tetu B. The molecular basis of Turcot’s syndrome. N Engl J Med. 1995;332:839-847. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 789] [Cited by in F6Publishing: 810] [Article Influence: 27.9] [Reference Citation Analysis (0)] |
| 33. | Furuuchi K, Tada M, Yamada H, Kataoka A, Furuuchi N, Hamada J, Takahashi M, Todo S, Moriuchi T. Somatic mutations of the APC gene in primary breast cancers. Am J Pathol. 2000;156:1997-2005. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 78] [Cited by in F6Publishing: 87] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 34. | Barth AI, Caro-Gonzalez HY, Nelson WJ. Role of adenomatous polyposis coli (APC) and microtubules in directional cell migration and neuronal polarization. Semin Cell Dev Biol. 2008;19:245-251. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 65] [Cited by in F6Publishing: 72] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 35. | Zhang L, Shay JW. Multiple Roles of APC and its Therapeutic Implications in Colorectal Cancer. J Natl Cancer Inst. 2017;109. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 159] [Cited by in F6Publishing: 221] [Article Influence: 31.6] [Reference Citation Analysis (0)] |
| 36. | Munemitsu S, Souza B, Müller O, Albert I, Rubinfeld B, Polakis P. The APC gene product associates with microtubules in vivo and promotes their assembly in vitro. Cancer Res. 1994;54:3676-3681. [PubMed] [Cited in This Article: ] |
| 37. | Zumbrunn J, Kinoshita K, Hyman AA, Näthke IS. Binding of the adenomatous polyposis coli protein to microtubules increases microtubule stability and is regulated by GSK3 beta phosphorylation. Curr Biol. 2001;11:44-49. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 331] [Cited by in F6Publishing: 361] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 38. | Mogensen MM, Tucker JB, Mackie JB, Prescott AR, Näthke IS. The adenomatous polyposis coli protein unambiguously localizes to microtubule plus ends and is involved in establishing parallel arrays of microtubule bundles in highly polarized epithelial cells. J Cell Biol. 2002;157:1041-1048. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 119] [Cited by in F6Publishing: 125] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 39. | Bellis J, Duluc I, Romagnolo B, Perret C, Faux MC, Dujardin D, Formstone C, Lightowler S, Ramsay RG, Freund JN. The tumor suppressor Apc controls planar cell polarities central to gut homeostasis. J Cell Biol. 2012;198:331-341. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 29] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 40. | Marshall TW, Lloyd IE, Delalande JM, Näthke I, Rosenblatt J. The tumor suppressor adenomatous polyposis coli controls the direction in which a cell extrudes from an epithelium. Mol Biol Cell. 2011;22:3962-3970. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 68] [Cited by in F6Publishing: 68] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 41. | Sandoval IT, Delacruz RG, Miller BN, Hill S, Olson KA, Gabriel AE, Boyd K, Satterfield C, Remmen HV, Rutter J. A metabolic switch controls intestinal differentiation downstream of Adenomatous polyposis coli (APC). Elife. 2017;6. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 21] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 42. | Nadauld LD, Phelps R, Moore BC, Eisinger A, Sandoval IT, Chidester S, Peterson PW, Manos EJ, Sklow B, Burt RW. Adenomatous polyposis coli control of C-terminal binding protein-1 stability regulates expression of intestinal retinol dehydrogenases. J Biol Chem. 2006;281:37828-37835. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 45] [Cited by in F6Publishing: 46] [Article Influence: 2.6] [Reference Citation Analysis (0)] |