Copyright
©The Author(s) 2023.
World J Clin Cases. Jul 16, 2023; 11(20): 4763-4787
Published online Jul 16, 2023. doi: 10.12998/wjcc.v11.i20.4763
Published online Jul 16, 2023. doi: 10.12998/wjcc.v11.i20.4763
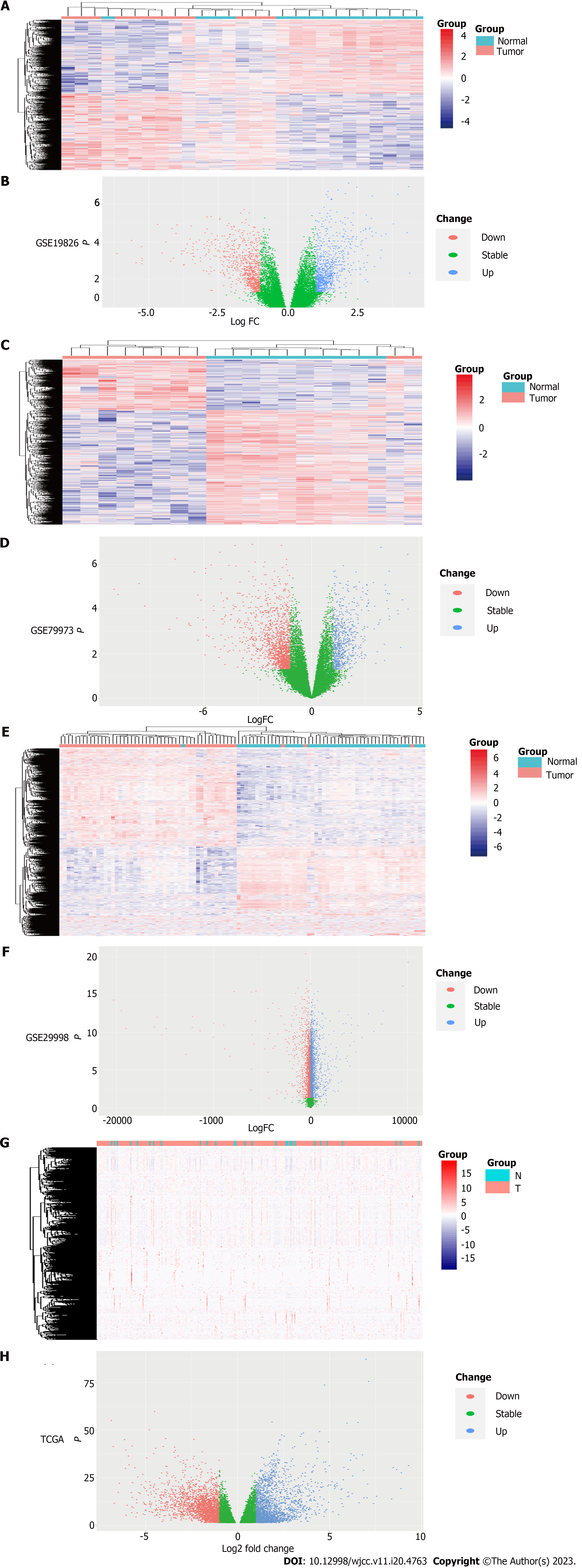
Figure 2 Identification of differentially expressed genes from The Cancer Genome Atlas and Gene Expression Omnibus Datasets.
A: The heat map of GSE19826 data set, with 2202 up-regulated genes and 2700 down-regulated genes. The blue normal samples, and the red are tumor samples; B: The volcanic map of GSE19826 dataset; C: The heat map of GSE79973 dataset, with 665 up-regulated genes and 1507 down-regulated genes. The blue are normal samples, and the red are tumor samples; D: The volcanic map of GSE79973 dataset; E: The heat map of GSE29998 dataset, with 4346 up-regulated genes and 3002 down-regulated genes. The blue are normal samples, and the red are tumor samples; F: The volcanic map of GSE29998 data set; G: The heat map of The Cancer Genome Atlas (TCGA) data set, with 2133 up-regulated genes and 2349 down-regulated genes. The blue are normal samples, and the red are tumor samples; H: The volcanic map of TCGA dataset. In B, D, F and H, the red dots are down-regulated genes and the blue dots are up-regulated genes. TCGA: The Cancer Genome Atlas.
- Citation: Yin LK, Yuan HY, Liu JJ, Xu XL, Wang W, Bai XY, Wang P. Identification of survival-associated biomarkers based on three datasets by bioinformatics analysis in gastric cancer. World J Clin Cases 2023; 11(20): 4763-4787
- URL: https://www.wjgnet.com/2307-8960/full/v11/i20/4763.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v11.i20.4763