Published online Jul 16, 2023. doi: 10.12998/wjcc.v11.i20.4788
Peer-review started: March 20, 2023
First decision: April 11, 2023
Revised: April 24, 2023
Accepted: June 13, 2023
Article in press: June 13, 2023
Published online: July 16, 2023
Breast cancer (BC) remains a public health problem. Tamoxifen (TAM) resistance has caused great difficulties for treatment of BC patients. Eukaryotic translation initiation factor 4E binding protein 1 (EIF4EBP1) plays critical roles in the tumorigenesis and progression of BC. However, the expression and mechanism of EIF4EBP1 in determining the efficacy of TAM therapy in BC patients are still unclear.
To investigate the expression and functions of EIF4EBP1 in determining the efficacy of TAM therapy in BC patients.
High-throughput sequencing data of breast tumors were downloaded from the Gene Expression Omnibus database. Differential gene expression analysis identified EIF4EBP1 to be significantly upregulated in cancer tissues. Its prognostic value was analyzed. The biological function and related pathways of EIF4EBP1 was analyzed. Subsequently, the expression of EIF4EBP1 was deter
EIF4EBP1 was upregulated in the TAM-resistant cells, and EIF4EBP1 was related to the prognosis of BC patients. Gene Set Enrichment Analysis showed that EIF4EBP1 might be involved in Hedgehog signaling pathways. Decreasing the expression of EIF4EBP1 could reverse TAM resistance, whereas overexpression of EIF4EBP1 promoted TAM resistance.
This study indicated that EIF4EBP1 was overexpressed in the BC and TAM-resistant cell line, which increased cell proliferation, invasion, migration and TAM resistance in BC cells.
Core Tip: Breast cancer is the most frequently diagnosed cancer in women and the leading cause of female deaths from cancer worldwide. Eukaryotic translation initiation factor 4E binding protein 1 was overexpressed in the breast cancer tamoxifen-resistant cell line, and high expression of eukaryotic translation initiation factor 4E binding protein 1 was associated with poor prognosis in tamoxifen-treated patients.
- Citation: Yang S, Hui TL, Wang HQ, Zhang X, Mi YZ, Cheng M, Gao W, Geng CZ, Li SN. High expression of autophagy-related gene EIF4EBP1 could promote tamoxifen resistance and predict poor prognosis in breast cancer. World J Clin Cases 2023; 11(20): 4788-4799
- URL: https://www.wjgnet.com/2307-8960/full/v11/i20/4788.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v11.i20.4788
Breast cancer (BC) is the most frequently diagnosed cancer in females and the leading cause of female deaths from cancer worldwide. BC is the most prevalent cancer worldwide. An estimated 3.9 million women have been diagnosed with BC in the past 5 years, and more than 350000 annual deaths worldwide are attributed to BC[1,2]. Estrogen receptor alpha (ERα) is expressed in more than 70% of all BC cases, playing key roles in the gene transcription of genes related to the development of BC cells[3,4]. Endocrine therapy is the major treatment strategy for both premenopausal and postmenopausal ER-positive patients. It functions by blocking the ERs or inhibiting estrogen production[5,6].
Tamoxifen (TAM), a selective estrogen modulator, is the most frequently prescribed antiestrogenic medication in the BC setting[7]. The introduction of TAM has significantly prolonged the overall survival (OS) and disease-free survival of BC patients[8,9]. However, approximately half of BC patients have intrinsic resistance to TAM or develop acquired drug resistance to the medication during treatment. TAM resistance remains one of the major causes of BC mortality today[10]. Therefore, it is necessary to identify biomarkers and therapeutic targets and to understand the molecular mechanisms of TAM resistance to improve patient survival.
Autophagy is a lysosomal degradation process that plays critical roles in cell survival and maintenance through the degradation of cytoplasmic organelles, proteins and macromolecules as well as the recovery of metabolites[11,12]. Studies have shown that defects in autophagy pathways can promote or inhibit drug resistance in many cancer types[13,14]. Eukaryotic translation initiation factor 4E binding protein 1 (EIF4EBP1) encodes one member of a family of translational repressor proteins that directly interacts with eIF4E. The Human Autophagy Database (http://www.autophagy.lu/) provides a complete list of human genes and proteins that are directly or indirectly involved in autophagy, and EIF4EBP1 is found in this database.
Rutkovsky et al[15] found that EIF4EBP1 was overexpressed in BC cells and that knockdown of EIF4EBP1 led to dramatic reductions in cell growth. Du et al[16] showed that EIF4EBP1 has significant prognostic value for BC. It has been reported that EIF4EBP1 is an independent prognostic factor for progesterone receptor-positive BC and that high expression of EIF4EBP1 was associated with drug resistance to endocrine treatment in the ER/progesterone receptor-positive patients[17]. Moreover, Hsieh et al[18] illustrated that EIF4EBP1 could enhance drug resistance in prostate cancer cells. However, the expression and molecular mechanism of EIF4EBP1 in TAM resistance in BC remains unknown.
In this study, we investigated the effects of EIF4EBP1 on TAM resistance and established it as a novel biomarker for TAM resistance. Bioinformatics analysis indicated that EIF4EBP1 was overexpressed in TAM-resistant BC cells. Moreover, high expression of EIF4EBP1 was associated with poor prognosis in TAM-resistant BC patients with TAM resistance and with a higher probability of metastasis and endocrine therapy resistance. In vitro experiments indicated that the expression level of EIF4EBP1 was positively correlated with TAM resistance in TAM-resistant BC cells.
In this study, high-throughput sequencing data of TAM-resistant BC were downloaded from the Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) database. The GSE21648 dataset contains one TAM-sensitive sample and six TAM-resistant samples, and the GSE26459 dataset contains three TAM-sensitive samples and three TAM-resistant samples. Then, the data in these datasets were normalized and summarized by the R software packages “limma” and “affy.” The R software package “limma” was used to identify differentially expressed genes (DEGs) in these datasets. The Benjamini-Hochberg method was used to adjust the fold change and P values. For DEGs, the cut-off criteria were
Gene Expression Profiling Interactive Analysis (GEPIA, http://gepia.cancer-pku.cn/) is a web-based tool that can be used to conduct patient survival analysis based on The Cancer Genome Atlas database[19]. Kaplan-Meier plotter (https://kmplot.com/analysis/) is a web-based tool that can be used to conduct patient survival analysis based on the Gene Expression Omnibus, EGA, and The Cancer Genome Atlas databases[20]. In this study, survival was compared between the high and low EIF4EBP1 expression groups based on GEPIA and Kaplan-Meier plotter. The cutoff criterion was P < 0.05. GSEA is a powerful method to explore biological insights and potential pathways related to a gene list by determining the genes that were in previously identified gene sets[21]. In this study, GSEA was used to explore the potential functions and molecular mechanisms of EIF4EBP1 in TAM-resistant BC cells. The six TAM-resistant BC samples in GSE21648 were classified into two groups according to the median expression level of EIF4EBP1. Then, GSEA was conducted by the R package “clusterProfiler”. The reference gene set used for GSEA was h.all.v6.2.sytmbols.gmt obtained from the Molecular Signatures Database.
There were 71 BC tissue specimens acquired from patients at the Fourth Hospital of Hebei Medical University (Shijiazhuang, China). The diagnosis of BC was made by two pathologists, and none of the patients had received any treatment (radiotherapy or endocrine therapy). This study was approved by the Ethics Committee of the Fourth Hospital of Hebei Medical University. All individuals were informed of the purpose of the study and had given written informed consent.
The BC cell line T47D (ER-positive) was purchased from the American Type Culture Collection (Manassas, VA, United States). 4-hydroxytamoxifen was purchased from Sigma-Aldrich (Shanghai, China). The TAM-resistant T47D-R cell line was obtained by continuous exposure to 4-hydroxytamoxifen (6 μM). Cells were maintained in a humidified incubator with 5% CO2 at 37 °C.
The expression of EIF4EBP1 in BC samples and cell culture was measured by real-time reverse transcription polymerase chain reaction (RT-qPCR). RNAiso Plus (TaKaRa, Otsu, Japan) was used to extract total RNA from BC cells. The PrimeScriptTM RT Reagent Kit (TaKaRa) was used to generate complementary DNA. RT-qPCR was performed using the TB Green Premix Ex Taq™ II kit on a MasterCycler5333 instrument (Eppendorf, Hamburg, Germany) according to the manufacturer’s instructions. The 2ΔΔCq method was used to calculate the relative expression of genes. GAPDH was used as an internal control.
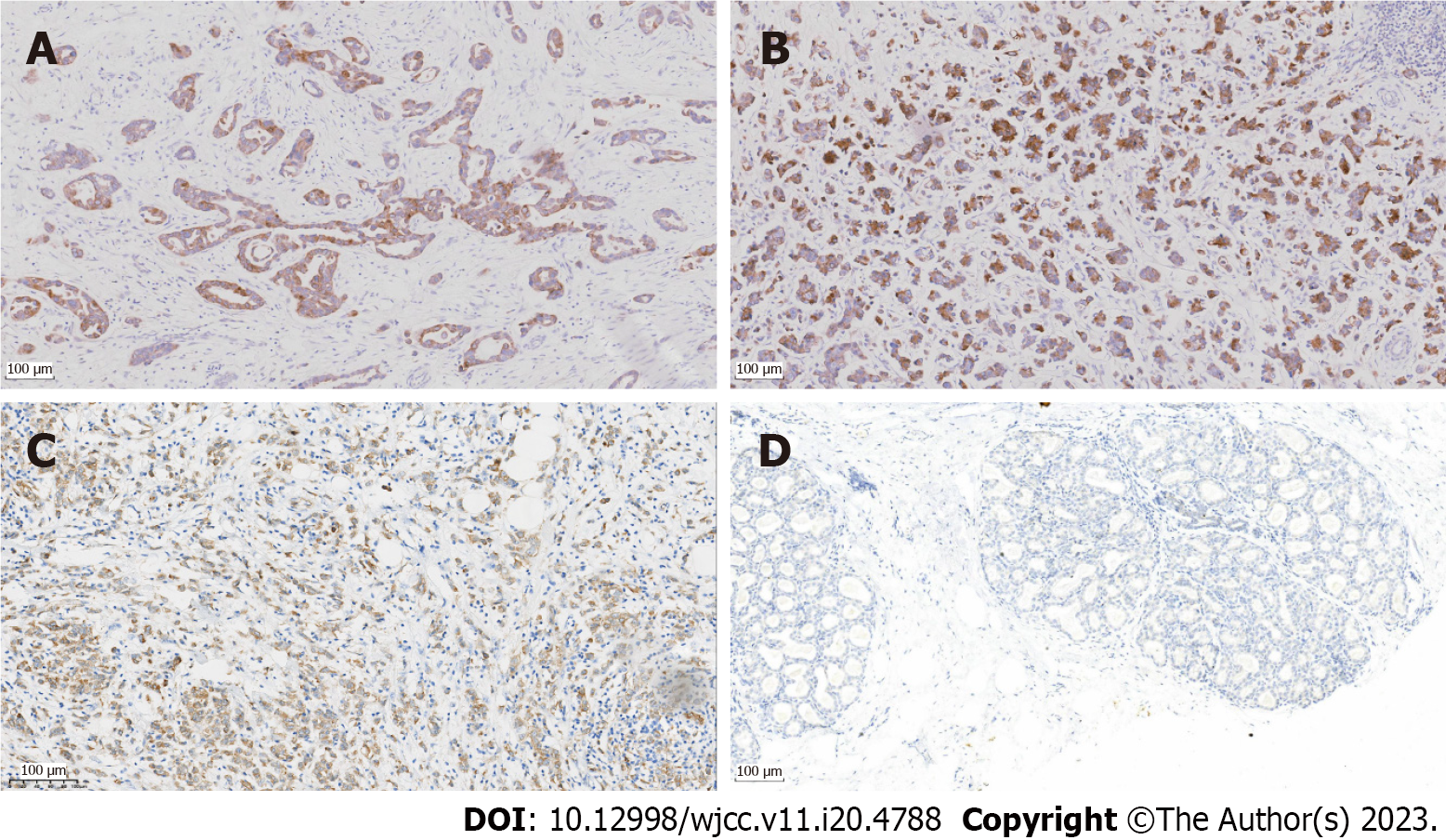
In this study, the expression of EIF4EBP1 in BC tissues from our hospital was determined by immunohistochemistry assay. Immunohistochemical analysis of BC samples were performed according to standard protocols. In brief, paraffin sections were dewaxed using xylene and rehydrated in a graded ethanol series. Hydrogen peroxide (0.3%) was used to block endogenous peroxidase activity. The Ventana Discovery XT automated stainer was used for immunohistochemistry, and ImageJ software was used for visualization.
Short interference RNAs (siRNAs) for EIF4EBP1 and corresponding scrambled siRNA negative controls were synthesized by GenePharma (Shanghai, China). The EIF4EBP1 plasmid and negative control vector were purchased from GenePharma (Shanghai, China). Lipofectamine 3000 transfection reagent (Thermo Fisher Scientific) was used to transiently transfect cells according to the manufacturer’s instructions. The transfection efficiency was evaluated by RT-qPCR after 24 h. Then, the slides with tissue samples were heated in sodium citrate buffer (95 °C, 10 min) and sealed in normal goat serum (37 °C, 1 h). The tissue samples were incubated with an anti-EIF4EBP1 antibody (Abcam, Shanghai, China) for 1 h at 37 °C. Then, the tissue samples were incubated with a secondary antibody conjugated with horseradish peroxidase (Abcam). The experimental results were independently assessed by three blinded pathologists and analyzed using ImageProPlus software (version 6; MediaCybernetics, Rockville, MD, United States).
A Cell Counting Kit-8 (CCK-8) assay was performed according to the manufacturer’s instructions to evaluate the effect of EIF4EBP1 knockdown or overexpression of EIF4EBP1 on the sensitivity of T47D-R cells to TAM treatment. T47D-R cells were seeded into 96-well plates (5000 cells/well) and incubated with TAM at different concentrations for 48 h. Subsequently, 10 μL of CCK-8 solution (Abcam) was added to each well. The OD450 values were measured with a microplate reader. Experiments were performed in triplicate.
To evaluate the effect of EIF4EBP1 knockdown or overexpression of EIF4EBP1 on the proliferation of the T47D-R cells after TAM treatment, a colony formation assay was performed. Cells in the logarithmic growth phase were seeded in 6-well plates at a density of 400 cells/well. T47D-R cells were cultured in a medium supplemented with 5 μM TAM. After 2 wk, cell colonies were fixed with 4% paraformaldehyde and visualized by staining with 0.1% crystal violet. Then cell colonies were counted and photographed. Experiments were performed in triplicate.
The invasion and migration abilities of T47D-R cells with knockdown or overexpression of EIF4EBP1 were analyzed via a transwell assay. T47D-R cells were seeded into 6-well plates at 1 × 105 cells/well for 24 h. Then T47D-R cells (200 mL/well) were seeded in a transwell chamber with 10% TBS and culture medium. The cells were cultured at 37 °C with 5% CO2 for 24 h. Then the liquid in the transwell chamber was removed. Cells in the lower chamber were fixed with 100% methanol, stained with 0.1% crystal violet and observed under a microscope. Experiments were performed in triplicate.
The invasion and migration abilities of T47D-R cells with EIF4EBP1 knockdown or overexpression of EIF4EBP1 were analyzed by wound healing assay. T47D-R cells were seeded in 6-well culture plates at 1 × 105 cells/well. After overnight incubation with 5 μM TAM, a wound was created with a sterile pipette tip. Then, the cells were imaged at 0 h and 48 h after the wound was created. Experiments were performed in triplicate.
In this study, all statistical analyses were performed by R software (version 3.5.3), SPSS 22.0 (SPSS Inc., Chicago, IL, United States) and GraphPad Prism 7.0. The R software package “limma” was used to identify DEGs. The OS of patients was analyzed by the R software package “survival ROC”. The experimental data were presented as the mean ± standard deviation values. The differences between the low and high EIF4EBP1 expression groups were compared by Pearson’s χ2 test. The Kaplan-Meier method was employed to analyze the OS of BC patients, and the log-rank test was used to estimate the differences between groups. P values less than 0.05 were considered to be statistically significant.
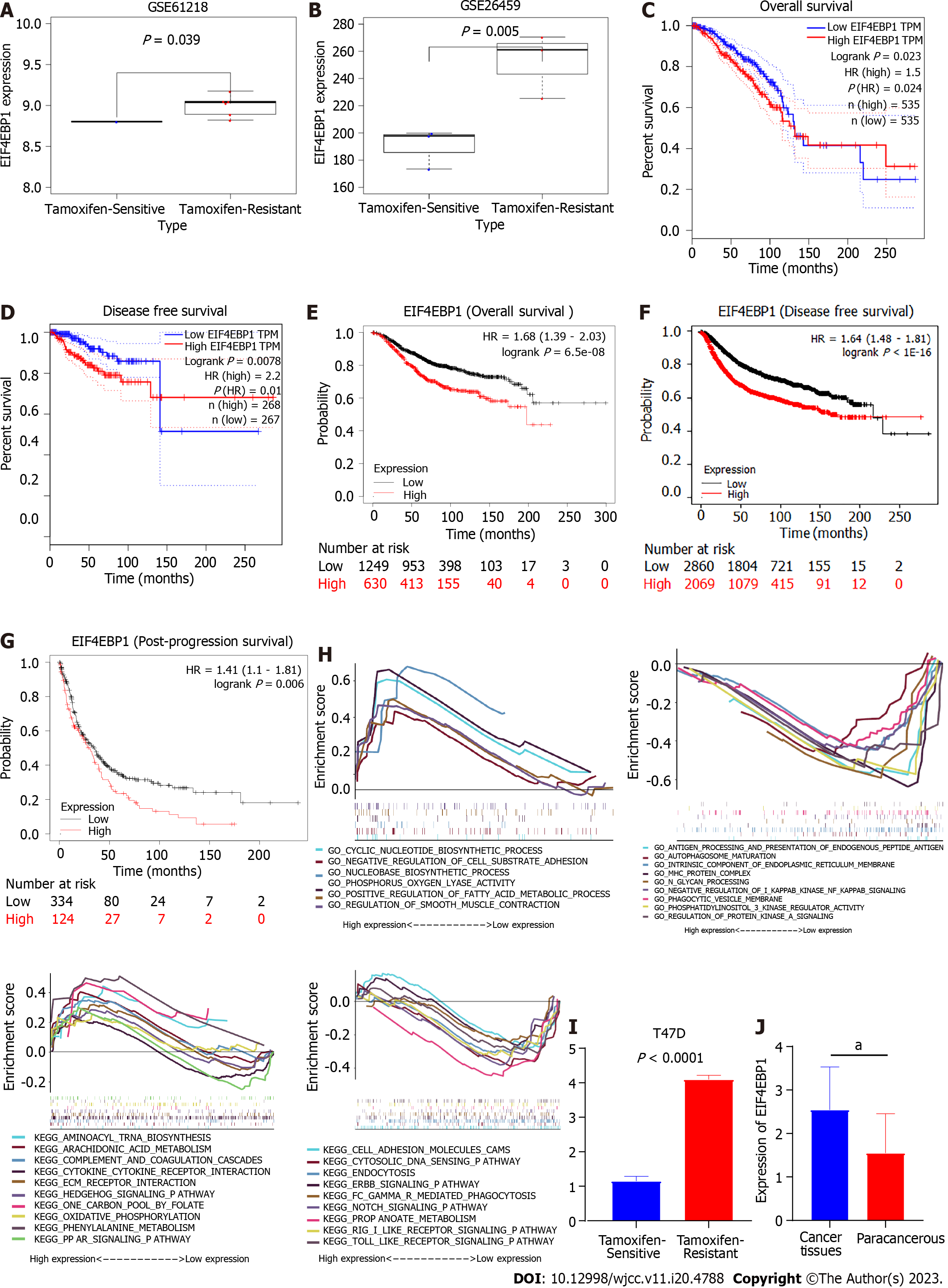
In this study, high-throughput sequencing data of TAM-resistant BC cells were downloaded and re-analyzed. As shown in Figures 1A and B, EIF4EBP1 was upregulated in TAM-resistant BC cells in GSE21618 and GSE26459. Survival analysis based on the GEPIA database suggested that high expression of EIF4EBP1 was significantly associated with worse OS (P = 0.023, Figure 1C) and disease-free survival (P = 0.01, Figure 1D). Moreover, survival analysis based on the Kaplan-Meier plotter database showed that high expression of EIF4EBP1 was significantly associated with worse OS (P = 6.5e-08, Figure 1E), disease-free survival (P < 1E-16, Figure 1F) and post-progression survival (P = 6.5e-08, Figure 1G). These results suggested that EIF4EBP1 could be a prognostic marker for BC patients.
The potential functions and molecular mechanisms of EIF4EBP1 in TAM-resistant BC cells were explored by GSEA. The TAM-resistant BC samples in GSE21618 were divided into two groups according to the median expression level of EIF4EBP1. Then, the potential functions and molecular mechanisms were explored by GSEA. Gene Ontology analysis based on the GSEA results suggested that cyclic nucleotide biosynthetic processes, negative regulation of cell substrate adhesion and nucleobase biosynthetic processes were upregulated, whereas antigen processing and presentation of endogenous peptide antigen, autophagosome maturation and intrinsic component of endoplasmic reticulum membrane were downregulated (Figure 1H). Kyoto Encyclopedia of Genes and Genomes analysis indicated that aminoacyl tRNA biosynthesis, the Hedgehog (Hh) signaling pathway and the peroxisome proliferator-activated receptor (PPAR) signaling pathway were upregulated, while the cell adhesion molecules cams, cytosolic DNA sensing pathways and ErbB signaling pathways were downregulated (Figure 1H).
In this study, RT-qPCR and immunohistochemistry were used to explore the expression of EIF4EBP1. As shown in Figures 1I, 1J and 2, EIF4EBP1 was significantly upregulated in BC tissues and TAM-resistant T47D cells. Moreover, the expression of EIF4EBP1 in BC tissues obtained at our hospital was determined, and the correlations of its expression with clinicopathological data (age, lymph node metastasis, radiotherapy status, endocrine therapy status, tumor stage, histological grade and metastasis stage) were calculated (Table 1). As shown in Table 1, the expression of EIF4EBP1 was significantly associated with lymph node metastasis (P < 0.0001), endocrine therapy status (P = 0.0005) and metastasis stage (P < 0.0001).
| Groups | n | Expression of EIF4EBP1 | P value | |
| Low expression | High expression | |||
| Age in yr | 0.5273 | |||
| < 50 | 34 | 20 | 14 | |
| ≥ 50 | 37 | 19 | 18 | |
| Metastasis of lymph nodes | 0.0000 | |||
| Negative | 23 | 20 | 3 | |
| 1 ≤ N+ ≤ 3 | 20 | 12 | 8 | |
| N+ > 3 | 28 | 7 | 21 | |
| Radiotherapy | 0.7274 | |||
| No | 36 | 20 | 16 | |
| Yes | 35 | 18 | 17 | |
| Chemotherapy | 0.0005 | |||
| TAM | 52 | 35 | 17 | |
| TAM + OFS | 19 | 4 | 15 | |
| Tumor stage | 0.8371 | |||
| I | 17 | 9 | 8 | |
| II | 44 | 24 | 20 | |
| III | 9 | 5 | 4 | |
| IV | 1 | 1 | 0 | |
| Histological grade | 0.3111 | |||
| I-II | 59 | 34 | 25 | |
| III | 12 | 5 | 7 | |
| Metastasis stage | 0.0000 | |||
| M0 | 48 | 35 | 13 | |
| M1 | 23 | 4 | 19 | |
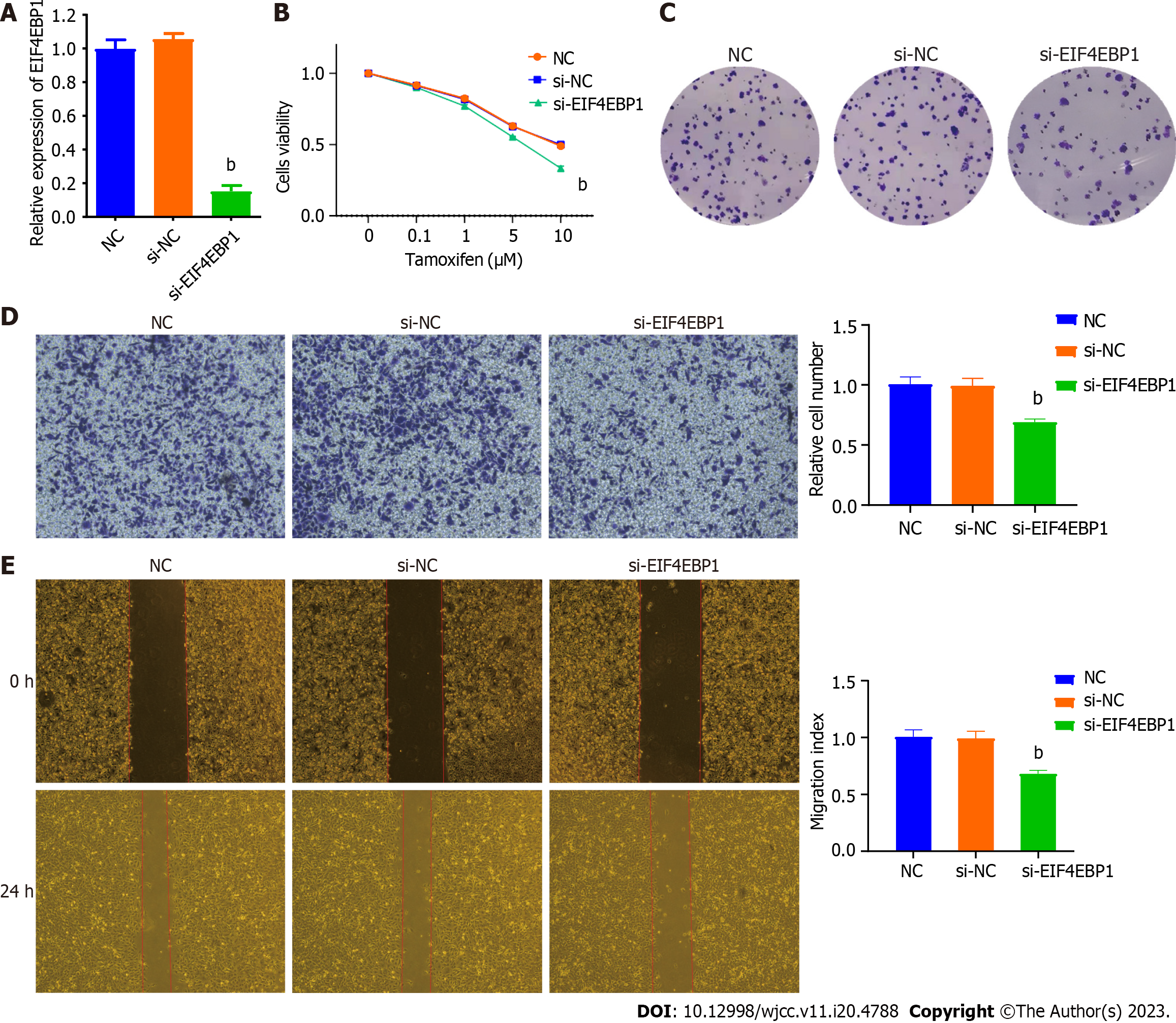
To explore the functions of EIF4EBP1 in TAM-resistant cells, the expression of EIF4EBP1 was reduced by transfecting siRNAs targeting EIF4EBP1. RT-qPCR showed that transient transfection of siRNA significantly decreased the expression of EIF4EBP1 (Figure 3A). The results of a CCK-8 assay suggested that downregulation of EIF4EBP1 significantly decreased the degree of resensitization to TAM in T47D-R cells (Figure 3B). Colony formation experiments indicated that the number of colonies consisting of T47D-R cells was significantly decreased after siRNA transfection (Figure 3C). The results of the transwell and wound healing assays indicated that EIF4EBP1 knockdown could reduce the invasion and migration of T47D-R cells treated with TAM. These results indicated that knockdown of EIF4EBP1 caused T47D-R cells to be resensitized to TAM.
To further understand the functions of EIF4EBP1 in TAM-resistant cells, the expression of EIF4EBP1 was upregulated by the transient transfection of a plasmid expressing EIF4EBP1. The overexpression of EIF4EBP1 was confirmed by RT-qPCR (Figure 4A). The cell viability of T47D-R cells treated with TAM was assessed by CCK-8 and colony formation assays. The results indicated that the resistance of T47D-R cells was further enhanced after EIF4EBP1 plasmid transfection (Figures 4B and C). The invasion and migration of T47D-R cells treated with TAM were explored by transwell and wound healing assays. The results suggested that cell invasion and migration were increased by EIF4EBP1 overexpression. These results indicate that the overexpression of EIF4EBP1 could increase the resistance of T47D-R cells to TAM.
BC is one of the most common cancers in the world, and ER-positive BC is the most common subtype of BC. Endocrine therapy, which targets the ER directly and/or suppresses estrogen production, is the main treatment strategy for ER-positive BC[21]. TAM is a nonsteroidal antiestrogen drug and has historically been the most widely used antiestrogen drug for the treatment of ER-positive BC patients[19,20]. Although TAM has greatly reduced the recurrence and mortality of BC, the emerging and acquired resistance to TAM has been a major obstacle for the successful treatment of patients[22,23]. Many studies have been conducted on the potential mechanism of TAM resistance, and several mechanisms have been shown to be related to TAM resistance. These mechanisms include autophagy, mutations of the ER and endoplasmic reticulum stress[10,22,24,25].
Autophagy is a “self-degradative” process in which cellular materials are sent to lysosomes for degradation. Autophagy plays a critical role in the turnover of cell components and provides energy and macromolecules[26,27]. Studies have shown that autophagy plays dual, context-dependent roles in drug resistance: It can kill drug-resistant cancer cells with inactive apoptotic pathways, but it can also participate in the development of drug resistance and protect cancer cells from endocrine therapy drugs[13,14].
EIF4EBP1 directly interacts with a limiting component of the multisubunit complex that recruits 40S ribosomal subunits to the 5’ end of mRNAs. Further studies showed that EIF4EBP1 was upregulated in many cancer types including BC, hepatocellular carcinoma, lung squamous cell carcinoma and glioblastoma[16,28-30], suggesting that EIF4EBP1 plays an important role in tumorigenesis. Ito et al[31] showed that EIF4EBP1 was overexpressed and phosphorylated in renal cell carcinoma and was involved in the clinical chemoresistance of renal cell carcinoma cells to mechanistic target of rapamycin complex 1 inhibitors. Tsai et al[32] found that EIF4EBP1 could induce glioma stem-like cells through the epidermal growth factor receptor/protein kinase B cascade, making a major contribution to drug resistance. However, the expression and molecular mechanisms of EIF4EBP1 in TAM resistance in BC remained unrevealed.
In this study, we investigated the role of EIF4EBP1 in TAM resistance. Scholars have performed studies on the role of EIF4EBP1 in TAM resistance, and their conclusions of these studies were similar with no controversy. Du et al[16] pointed out in a 2020 study that EIF4EBP1 showed significant prognostic value as a prognostic indicator in BC, specifically indicating poor prognosis. A 2019 study reported that EIF4EBP1 was located within the 8p11-p12 genomic locus, frequently highly amplified in BC and predicted poor prognosis and resistance to endocrine therapy. Another study, from 2022, indicated that the addition of EIF4EBP1 to cultures significantly reduced the proliferation and metastasis of TNBC cells[33]. These studies demonstrated that EIF4EBP1 plays an oncogenic role in BC.
However, most of these conclusions are based on bioinformatics analysis. The role of EIF4EBP1 in TAM resistance has not been experimentally demonstrated. This study indicated that EIF4EBP1 enhanced the resistance of T47D-R cells to TAM. EIF4EBP1 is an autophagy-related gene; some studies have demonstrated a role for EIF4EBP1 in autophagy. For example, it has been reported that in CACO-2 cells exposed to cetuximab, EIF4EBP1 expression and autophagosome formation increased, and autophagy increased the efficacy of cetuximab in colorectal cancer[34]. Moreover, Lai et al[35] indicated that YXM110 is a new synthetic drugs that exhibits excellent anti-tumor activity in many cancer cells by mediating EIF4EBP1 depletion and regulating autophagy. In this study, we suggested that EIF4EBP1 may increase the resistance of T47D-R cells to TAM by regulating autophagy.
Given the critical role played by EIF4EBP1 in the development of drug resistance and BC, we explored whether EIF4EBP1 is involved in TAM resistance. In this study, the gene expression profiles of TAM-resistant or TAM-sensitive BC cells were reanalyzed. EIF4EBP1 was overexpressed in TAM-resistant cells, and high expression of EIF4EBP1 was associated with poor prognosis in BC patients. Based on the clinical specimens from our hospital, we found that high expression of EIF4EBP1 was associated with metastasis and endocrine therapy in BC patients. Moreover, cell experiments suggested that EIF4EBP1 deficiency could reverse TAM resistance, whereas overexpression of EIF4EBP1 increased TAM resistance.
In this study, the potential functions and molecular mechanisms of EIF4EBP1 in TAM-resistant BC cells were identified by GSEA. Notably, the Hh signaling pathway was significantly enriched in the high EIF4EBP1 expression group. It has been reported that Hh signaling pathway is involved in developmental processes in vertebrates and that abnormal activation of this pathway plays an important role in tumorigenesis and maintenance of multiple cancers[36]. Ren et al[37] indicated that tumor suppressor candidate 3 may improve the expression of CD133 and ABCC1 by activating the Hh signaling pathway, and inhibition of the Hh signaling pathway could reduce drug resistance of colorectal cancer cells. Zeng et al[36] demonstrated that the inhibition of the Hh signaling pathway could induce autophagy in chronic myeloid leukemia cells, and inhibiting autophagy and the Hh signaling pathway could reduce cell viability and induce apoptosis of imatinib-resistant chronic myeloid leukemia cells. Thus, we hypothesize that EIF4EBP1 could induce TAM resistance through Hh signaling pathway and autophagy. However, the results need to be examined by further investigations.
We also found that components of the PPAR signaling pathway were significantly enriched in the high EIF4EBP1 expression group. The PPAR signaling pathway has been linked to glucose and lipid metabolic disorders, endothelial function and inflammation. It has been reported that PPAR-γ is upregulated in glioma cells, which could regulate genes associated with apoptosis and multidrug resistance and increase intracellular accumulation of drugs[38]. Moreover, Bräutigam et al[39] showed that the death ligand TRAIL (TNF superfamily member 10) could sensitize tumor cells to cytostatic drugs without affecting normal tissues. The combinatorial treatment with PPAR-γ ligands and TRAIL has been shown to synergistically induce apoptosis in ovarian cancer cell lines. Thus, we hypothesize that the PPAR-γ agonists may be promising drugs for targeting drug-resistant cells. Moreover, proteins in the ErbB signaling pathway were significantly enriched in the low EIF4EBP1 expression group. Studies have shown that the abnormal activation of ErbB family members is involved in tumorigenesis and in the escape from anti-tumor immunity in many types of cancers[40].
Song et al[41] indicated that the host genes of the identified circular RNAs in platinum-based drug-resistant non-small cell lung cancer cells were involved in the ErbB signaling pathway. Moreover, Macleod et al[42] indicated that ErbB receptor signaling was altered in cisplatin-resistant ovarian cancer cells, suggesting that downregulation of the ErbB signaling pathway could play an important role in the development of drug resistance. Thus, we hypothesize that EIF4EBP1 could induce TAM resistance by regulating the ErbB signaling pathway.
This study had some limitations. First, as with many previous studies[43,44], only T47D cells were used to establish TAM-resistant cell lines. This may make our conclusions less generalizable. Moreover, gene set enrichment analysis was used to explore the potential functions and molecular mechanisms of EIF4EBP1 in TAM-resistant BC cells. However, these pathways have not been verified by in vitro and in vivo experiments.
In conclusion, our study showed that the overexpression of EIF4EBP1 was significantly associated with poor prognosis and metastasis in BC patients. Moreover, EIF4EBP1 plays important roles in the development of TAM resistance. EIF4EBP1 knockdown could reverse TAM resistance, whereas overexpression of EIF4EBP1 increased TAM resistance in BC cells. In addition, our GSEA results may provide new insights into the molecular mechanism of TAM resistance. In brief, EIF4EBP1 could be a marker for the early diagnosis and a therapeutic target for the therapy of TAM resistance.
Tamoxifen (TAM) resistance is a major obstacle in the treatment of breast cancer (BC) patients. It has been reported that eukaryotic translation initiation factor 4E binding protein 1 (EIF4EBP1) plays critical roles in the tumorigenesis and development of BC.
TAM resistance remains one of the major causes of BC mortality today. Therefore, it is necessary to identify biomarkers and therapeutic targets and understand molecular mechanisms of TAM resistance to help patients.
The objective was to investigate the expression and functions of EIF4EBP1 in the efficacy of TAM therapy in BC patients.
Gene Set Enrichment Analysis (GSEA) was performed to explore the biological functions and related pathways of EIF4EBP1. Real-time reverse transcription polymerase chain reaction were employed to explore the expression of EIF4EBP1 in TAM-resistant and TAM-sensitive BC cell lines. Cell count kit-8 assay, colony formation experiments and the wound healing assay were used to understand the phenotypes of loss- and gain-of-function of EIF4EBP1 in a TAM-resistant cell line.
EIF4EBP1 was upregulated in TAM resistant cells, and EIF4EBP1 was associated with the prognosis of BC patients. GSEA suggested that EIF4EBP1 may be involved in the Hedgehog signaling pathway. Reducing the expression of EIF4EBP1 can reverse TAM resistance, while overexpression of EIF4EBP2 can promote TAM resistance.
In this study, we investigated the role of EIF4EBP1 in TAM resistance. Scholars have performed studies on the role of EIF4EBP1 in TAM resistance, and their conclusions of these studies were similar with no controversy. However, most of these conclusions were based on bioinformatics analysis. The role of EIF4EBP1 in TAM resistance has not been experimentally demonstrated. This study indicated that EIF4EBP1 enhanced the resistance of T47D-R cells to TAM. In addition, our GSEA results may provide new insights into the molecular mechanism of TAM resistance. In brief, EIF4EBP1 could be a marker for the early diagnosis and a therapeutic target for the therapy of TAM resistance.
Further studies should explore the potential function and molecular mechanism of EIF4EBP1 in TAM-resistant BC cells through in vitro and in vivo experiments.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Medicine, research and experimental
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): E
P-Reviewer: Eid N, Malaysia; Emran TB, Bangladesh; Wong WL, China S-Editor: Wang JJ L-Editor: Filipodia P-Editor: Zhao S
| 1. | Parkin DM. Global cancer statistics in the year 2000. Lancet Oncol. 2001;2:533-543. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1727] [Cited by in F6Publishing: 1679] [Article Influence: 73.0] [Reference Citation Analysis (0)] |
| 2. | Zwirner J, Ondruschka B, Scholze M, Schulze-Tanzil G, Hammer N. Mechanical and morphological description of human acellular dura mater as a scaffold for surgical reconstruction. J Mech Behav Biomed Mater. 2019;96:38-44. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 3. | Siersbæk R, Kumar S, Carroll JS. Signaling pathways and steroid receptors modulating estrogen receptor α function in breast cancer. Genes Dev. 2018;32:1141-1154. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 99] [Cited by in F6Publishing: 90] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 4. | Sharma D, Kumar S, Narasimhan B. Estrogen alpha receptor antagonists for the treatment of breast cancer: a review. Chem Cent J. 2018;12:107. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 60] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 5. | Reinbolt RE, Mangini N, Hill JL, Levine LB, Dempsey JL, Singaravelu J, Koehler KA, Talley A, Lustberg MB. Endocrine therapy in breast cancer: the neoadjuvant, adjuvant, and metastatic approach. Semin Oncol Nurs. 2015;31:146-155. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 24] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 6. | Harbeck N, Gnant M. Breast cancer. Lancet. 2017;389:1134-1150. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1135] [Cited by in F6Publishing: 1361] [Article Influence: 194.4] [Reference Citation Analysis (0)] |
| 7. | Shagufta, Ahmad I. Tamoxifen a pioneering drug: An update on the therapeutic potential of tamoxifen derivatives. Eur J Med Chem. 2018;143:515-531. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 112] [Cited by in F6Publishing: 99] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 8. | Ruhstaller T, Giobbie-Hurder A, Colleoni M, Jensen MB, Ejlertsen B, de Azambuja E, Neven P, Láng I, Jakobsen EH, Gladieff L, Bonnefoi H, Harvey VJ, Spazzapan S, Tondini C, Del Mastro L, Veyret C, Simoncini E, Gianni L, Rochlitz C, Kralidis E, Zaman K, Jassem J, Piccart-Gebhart M, Di Leo A, Gelber RD, Coates AS, Goldhirsch A, Thürlimann B, Regan MM; members of the BIG 1-98 Collaborative Group and the International Breast Cancer Study Group. Adjuvant Letrozole and Tamoxifen Alone or Sequentially for Postmenopausal Women With Hormone Receptor-Positive Breast Cancer: Long-Term Follow-Up of the BIG 1-98 Trial. J Clin Oncol. 2019;37:105-114. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 61] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 9. | Mayer EL, Dueck AC, Martin M, Rubovszky G, Burstein HJ, Bellet-Ezquerra M, Miller KD, Zdenkowski N, Winer EP, Pfeiler G, Goetz M, Ruiz-Borrego M, Anderson D, Nowecki Z, Loibl S, Moulder S, Ring A, Fitzal F, Traina T, Chan A, Rugo HS, Lemieux J, Henao F, Lyss A, Antolin Novoa S, Wolff AC, Vetter M, Egle D, Morris PG, Mamounas EP, Gil-Gil MJ, Prat A, Fohler H, Metzger Filho O, Schwarz M, DuFrane C, Fumagalli D, Theall KP, Lu DR, Bartlett CH, Koehler M, Fesl C, DeMichele A, Gnant M. Palbociclib with adjuvant endocrine therapy in early breast cancer (PALLAS): interim analysis of a multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2021;22:212-222. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 96] [Cited by in F6Publishing: 148] [Article Influence: 49.3] [Reference Citation Analysis (0)] |
| 10. | Yao J, Deng K, Huang J, Zeng R, Zuo J. Progress in the Understanding of the Mechanism of Tamoxifen Resistance in Breast Cancer. Front Pharmacol. 2020;11:592912. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 57] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 11. | Galluzzi L, Green DR. Autophagy-Independent Functions of the Autophagy Machinery. Cell. 2019;177:1682-1699. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 339] [Cited by in F6Publishing: 531] [Article Influence: 132.8] [Reference Citation Analysis (0)] |
| 12. | Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27-42. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4916] [Cited by in F6Publishing: 5442] [Article Influence: 340.1] [Reference Citation Analysis (1)] |
| 13. | Smith AG, Macleod KF. Autophagy, cancer stem cells and drug resistance. J Pathol. 2019;247:708-718. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 160] [Cited by in F6Publishing: 236] [Article Influence: 47.2] [Reference Citation Analysis (0)] |
| 14. | Huang F, Wang BR, Wang YG. Role of autophagy in tumorigenesis, metastasis, targeted therapy and drug resistance of hepatocellular carcinoma. World J Gastroenterol. 2018;24:4643-4651. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 119] [Cited by in F6Publishing: 135] [Article Influence: 22.5] [Reference Citation Analysis (2)] |
| 15. | Rutkovsky AC, Yeh ES, Guest ST, Findlay VJ, Muise-Helmericks RC, Armeson K, Ethier SP. Eukaryotic initiation factor 4E-binding protein as an oncogene in breast cancer. BMC Cancer. 2019;19:491. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 33] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 16. | Du JX, Chen C, Luo YH, Cai JL, Cai CZ, Xu J, Ni XJ, Zhu W. Establishment and validation of a novel autophagy-related gene signature for patients with breast cancer. Gene. 2020;762:144974. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 17. | Karlsson E, Pérez-Tenorio G, Amin R, Bostner J, Skoog L, Fornander T, Sgroi DC, Nordenskjöld B, Hallbeck AL, Stål O. The mTOR effectors 4EBP1 and S6K2 are frequently coexpressed, and associated with a poor prognosis and endocrine resistance in breast cancer: a retrospective study including patients from the randomised Stockholm tamoxifen trials. Breast Cancer Res. 2013;15:R96. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 66] [Cited by in F6Publishing: 73] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 18. | Hsieh AC, Nguyen HG, Wen L, Edlind MP, Carroll PR, Kim W, Ruggero D. Cell type-specific abundance of 4EBP1 primes prostate cancer sensitivity or resistance to PI3K pathway inhibitors. Sci Signal. 2015;8:ra116. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 32] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 19. | Eggemann H, Bernreiter AL, Reinisch M, Loibl S, Taran FA, Costa SD, Ignatov A. Tamoxifen treatment for male breast cancer and risk of thromboembolism: prospective cohort analysis. Br J Cancer. 2019;120:301-305. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in F6Publishing: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 20. | Höner Zu Siederdissen C, Maasoumy B, Marra F, Deterding K, Port K, Manns MP, Cornberg M, Back D, Wedemeyer H. Drug-Drug Interactions With Novel All Oral Interferon-Free Antiviral Agents in a Large Real-World Cohort. Clin Infect Dis. 2016;62:561-567. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 78] [Cited by in F6Publishing: 83] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 21. | Hanker AB, Sudhan DR, Arteaga CL. Overcoming Endocrine Resistance in Breast Cancer. Cancer Cell. 2020;37:496-513. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 405] [Cited by in F6Publishing: 374] [Article Influence: 93.5] [Reference Citation Analysis (0)] |
| 22. | Clarke R, Tyson JJ, Dixon JM. Endocrine resistance in breast cancer--An overview and update. Mol Cell Endocrinol. 2015;418 Pt 3:220-234. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 228] [Cited by in F6Publishing: 224] [Article Influence: 24.9] [Reference Citation Analysis (0)] |
| 23. | Zhang J, Wang Q, Cao J, Sun J, Zhu Z. Mechanisms of resistance to estrogen receptor modulators in ER+/HER2- advanced breast cancer. Cell Mol Life Sci. 2020;77:559-572. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 24. | Cook KL, Shajahan AN, Clarke R. Autophagy and endocrine resistance in breast cancer. Expert Rev Anticancer Ther. 2011;11:1283-1294. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 112] [Cited by in F6Publishing: 120] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 25. | Tsoi H, You CP, Leung MH, Man EPS, Khoo US. Targeting Ribosome Biogenesis to Combat Tamoxifen Resistance in ER+ve Breast Cancer. Cancers (Basel). 2022;14. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2] [Cited by in F6Publishing: 8] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 26. | Levy JMM, Towers CG, Thorburn A. Targeting autophagy in cancer. Nat Rev Cancer. 2017;17:528-542. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1382] [Cited by in F6Publishing: 1695] [Article Influence: 242.1] [Reference Citation Analysis (0)] |
| 27. | Mizushima N, Levine B. Autophagy in Human Diseases. N Engl J Med. 2020;383:1564-1576. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 420] [Cited by in F6Publishing: 554] [Article Influence: 138.5] [Reference Citation Analysis (0)] |
| 28. | Cha YL, Li PD, Yuan LJ, Zhang MY, Zhang YJ, Rao HL, Zhang HZ, Zheng XF, Wang HY. EIF4EBP1 overexpression is associated with poor survival and disease progression in patients with hepatocellular carcinoma. PLoS One. 2015;10:e0117493. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 21] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 29. | Zhu J, Wang M, Hu D. Development of an autophagy-related gene prognostic signature in lung adenocarcinoma and lung squamous cell carcinoma. PeerJ. 2020;8:e8288. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 34] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 30. | Fan QW, Nicolaides TP, Weiss WA. Inhibiting 4EBP1 in Glioblastoma. Clin Cancer Res. 2018;24:14-21. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 30] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 31. | Ito H, Ichiyanagi O, Naito S, Bilim VN, Tomita Y, Kato T, Nagaoka A, Tsuchiya N. GSK-3 directly regulates phospho-4EBP1 in renal cell carcinoma cell-line: an intrinsic subcellular mechanism for resistance to mTORC1 inhibition. BMC Cancer. 2016;16:393. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in F6Publishing: 29] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 32. | Tsai YT, Wu AC, Yang WB, Kao TJ, Chuang JY, Chang WC, Hsu TI. ANGPTL4 Induces TMZ Resistance of Glioblastoma by Promoting Cancer Stemness Enrichment via the EGFR/AKT/4E-BP1 Cascade. Int J Mol Sci. 2019;20. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 23] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 33. | Yan C, Liu Q, Jia R. Construction and Validation of a Prognostic Risk Model for Triple-Negative Breast Cancer Based on Autophagy-Related Genes. Front Oncol. 2022;12:829045. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2] [Cited by in F6Publishing: 12] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 34. | Guo GF, Wang YX, Zhang YJ, Chen XX, Lu JB, Wang HH, Jiang C, Qiu HQ, Xia LP. Predictive and prognostic implications of 4E-BP1, Beclin-1, and LC3 for cetuximab treatment combined with chemotherapy in advanced colorectal cancer with wild-type KRAS: Analysis from real-world data. World J Gastroenterol. 2019;25:1840-1853. [PubMed] [DOI] [Cited in This Article: ] [Cited by in CrossRef: 18] [Cited by in F6Publishing: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 35. | Lai CY, Pan SL, Yang XM, Chang LH, Chang YL, Yang PC, Lee KH, Teng CM. Depletion of 4E-BP1 and regulation of autophagy lead to YXM110-induced anticancer effects. Carcinogenesis. 2013;34:2050-2060. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in F6Publishing: 8] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 36. | Zeng X, Ju D. Hedgehog Signaling Pathway and Autophagy in Cancer. Int J Mol Sci. 2018;19. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 67] [Cited by in F6Publishing: 70] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 37. | Ren Y, Deng R, Cai R, Lu X, Luo Y, Wang Z, Zhu Y, Yin M, Ding Y, Lin J. TUSC3 induces drug resistance and cellular stemness via Hedgehog signaling pathway in colorectal cancer. Carcinogenesis. 2020;41:1755-1766. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 8] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 38. | Han S, Lv X, Wang Y, Gong H, Zhang C, Tong A, Zhang B, Yao H. Effect and mechanism of peroxisome proliferator-activated receptor-γ on the drug resistance of the U-87 MG/CDDP human malignant glioma cell line. Mol Med Rep. 2015;12:2239-2246. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 9] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 39. | Bräutigam K, Biernath-Wüpping J, Bauerschlag DO, von Kaisenberg CS, Jonat W, Maass N, Arnold N, Meinhold-Heerlein I. Combined treatment with TRAIL and PPARγ ligands overcomes chemoresistance of ovarian cancer cell lines. J Cancer Res Clin Oncol. 2011;137:875-886. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 20] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 40. | Kumagai S, Koyama S, Nishikawa H. Antitumour immunity regulated by aberrant ERBB family signalling. Nat Rev Cancer. 2021;21:181-197. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 86] [Cited by in F6Publishing: 124] [Article Influence: 41.3] [Reference Citation Analysis (0)] |
| 41. | Song L, Cui Z, Guo X. Comprehensive analysis of circular RNA expression profiles in cisplatin-resistant non-small cell lung cancer cell lines. Acta Biochim Biophys Sin (Shanghai). 2020;52:944-953. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 42. | Macleod K, Mullen P, Sewell J, Rabiasz G, Lawrie S, Miller E, Smyth JF, Langdon SP. Altered ErbB receptor signaling and gene expression in cisplatin-resistant ovarian cancer. Cancer Res. 2005;65:6789-6800. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 109] [Cited by in F6Publishing: 115] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 43. | Gao A, Sun T, Ma G, Cao J, Hu Q, Chen L, Wang Y, Wang Q, Sun J, Wu R, Wu Q, Zhou J, Liu L, Hu J, Dong JT, Zhu Z. LEM4 confers tamoxifen resistance to breast cancer cells by activating cyclin D-CDK4/6-Rb and ERα pathway. Nat Commun. 2018;9:4180. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 41] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 44. | Zhou J, Li W, Ming J, Yang W, Lu L, Zhang Q, Ruan S, Huang T. High expression of TRAF4 predicts poor prognosis in tamoxifen-treated breast cancer and promotes tamoxifen resistance. Anticancer Drugs. 2020;31:558-566. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 5] [Article Influence: 1.7] [Reference Citation Analysis (0)] |