Published online Jun 6, 2023. doi: 10.12998/wjcc.v11.i16.3891
Peer-review started: March 14, 2023
First decision: March 28, 2023
Revised: April 6, 2023
Accepted: April 14, 2023
Article in press: April 14, 2023
Published online: June 6, 2023
Neurodevelopmental-craniofacial syndrome with variable renal and cardiac abnormalities (NECRC) is a rare, autosomal, dominant neurological disorder caused by mutations in the ZMYM2 gene. To date, the clinical and functional characteristics of the novel ZMYM2 mutation c.2090_2091del have not yet been reported.
The patient was an 18.5-mo-old Chinese boy with motor and language delay, microcephaly, facial dysmorphism, moderate malnutrition, single palmar crease on the left hand, synpolydactyly of the right foot, hypotonia and feeding pr
We performed a systematic literature review to identify and characterize NECRC. Substantial evidence from the literature indicated that patients with ZMYM2 gene mutation showed different degrees of intellectual disability, motor and language retardation, facial dysmorphism, and a few had congenital heart defects, kidney and urinary tract abnormalities. Early diagnosis and prompt management with comprehensive rehabilitation training are beneficial, but may not improve long-term outcomes.
Core Tip: We describe a patient with neurodevelopmental-craniofacial syndrome with variable renal and cardiac abnormalities caused by ZMYM2 mutation. Bioinformatics analysis suggested the presence of a novel complex heterozygous variant in the ZMYM2 gene.
- Citation: Li Y, Zhou Z, Xu Y, Wang ZR. Novel mutation c.2090_2091del in neurodevelopmental-craniofacial syndrome with variable renal and cardiac abnormalities in an 18.5-mo-old boy: A case report. World J Clin Cases 2023; 11(16): 3891-3898
- URL: https://www.wjgnet.com/2307-8960/full/v11/i16/3891.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v11.i16.3891
Neurodevelopmental-craniofacial syndrome with variable renal and cardiac abnormalities (NECRC) is an autosomal dominant disorder characterized by craniofacial dysmorphology associated with mild developmental delay, mildly impaired intellectual development or learning difficulties, speech delay, and behavioral abnormalities. Approximately half of patients have congenital abnormalities of the kidney and urinary tract (CAKUT) and/or congenital cardiac defects, including septal defects. We identified a patient with a novel de novo frameshift variant exhibiting the combinational phenotype of developmental delay and facial dysmorphism. Medication combined with comprehensive rehabilitation training was not effective. A literature review including the medical history, clinical symptoms and genetic features of NECRC patients with ZMYM2 mutations, which further specified the phenotype and genotype of these patients was also carried out. This case report was approved for publication by the Ethics Committees of the First Affiliated Hospital, Henan University of Chinese Medicine, and written informed consent was obtained from the patient’s parents and family.
The patient, an 18.5-mo-old boy, was admitted to the First Affiliated Hospital, Henan University of Chinese Medicine on August 31, 2022, due to developmental delay for more than 1 year.
Developmental delay, communication disability, attention deficit hyperactivity disorder, sluggish response, clumsy in movement and behavioral concerns were identified in this patient. He also exhibited decreased appetite, sleep deterioration, and ingestion of a liquid diet. Treatment with lysine hydrochloride resulted in a poor effect and zinc gluconate granules 35 mg twice daily on June 18, 2022 were ineffective. Oxiracetam capsules 400 mg twice daily were added on July 21, 2022 which were mildly effective.
At the age of 6 mo, the patient presented his initial symptom of motor retardation, he had a history of poor motor and language development milestones: he could not sit steadily until 12 mo, could walk with help at 17 mo, could not walk smoothly until 18.5 mo, and only had a single tone. The patient was diagnosed with "global developmental delay" aged 6.9 mo.
The patient had an abnormal birth history: His mother, a 29-year-old woman, was hospitalized for 2 wk due to small fetal heart at 35 wk of gestation. The boy was delivered by cesarean section at 37 wk of pregnancy because of oligohydramnios, his birth weight was 3000 g (-1 SD), birth length was 51.0 cm (0.80 SD) with no history of asphyxia or hypoxia. The boy is the second child of healthy non-consanguineous parents, his elder sister could only say "baba" and "mama" aged 2 years, and whose language is currently slightly delayed aged 4 years and 7 mo. Her intelligence, height, weight and facial features are normal. There is no family history of intellectual disability, motor or language retardation.
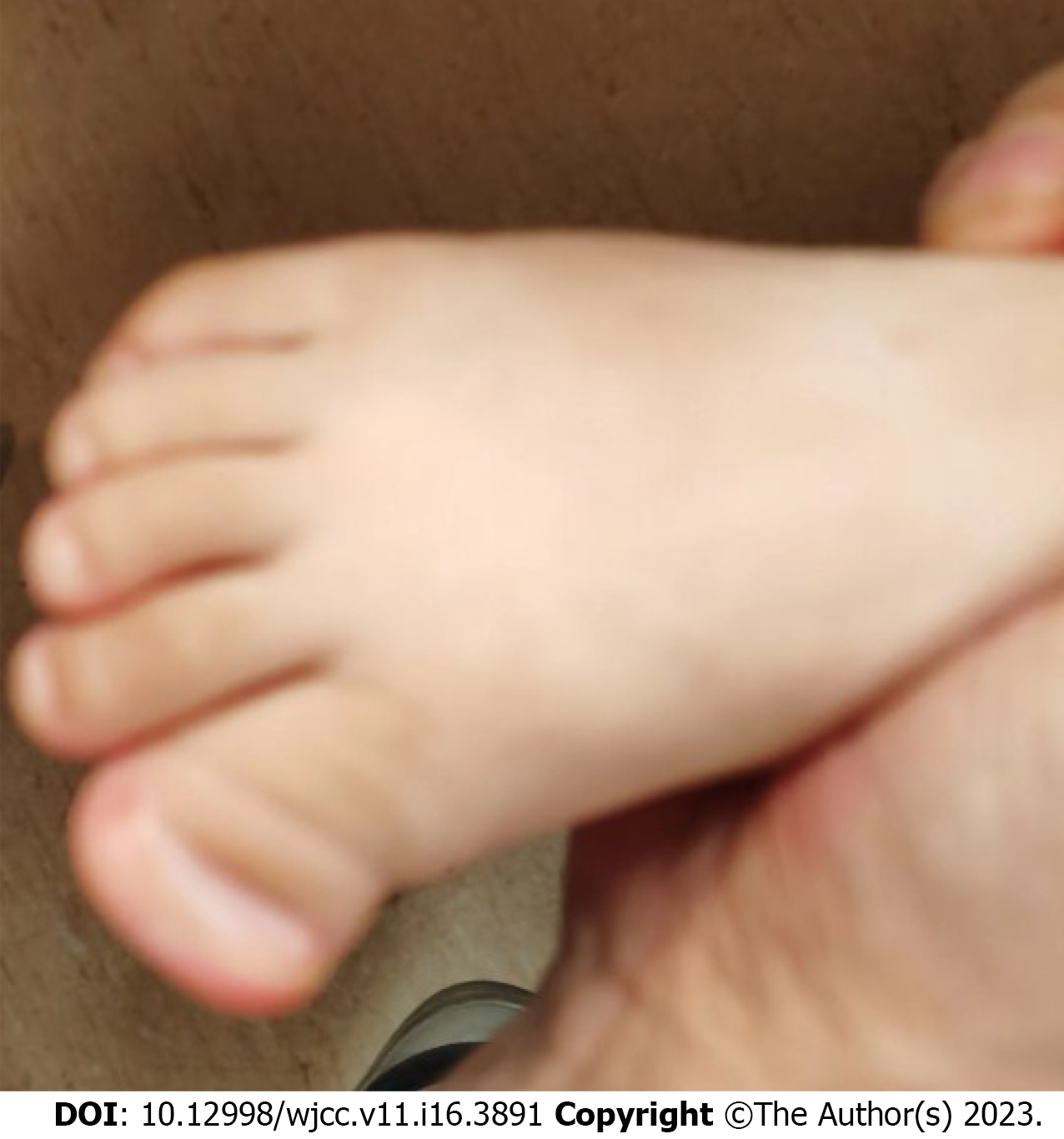
At 18.5 mo old, his height is 85.5 cm (0.50 SD), weight is 9.2 kg (-2 SD), and head circumference is 45.5 cm (-0.5 SD). He has protruding ears, wide interpupillary distance, broad nasal bridge, thin lips, single palmar crease on the left hand, and synpolydactyly of the right foot (Figure 1). No obvious abnor
The child has performed the Gesell Developmental Schedule (GDS) four times at different ages (Table 1). At 18.5 mo, the Alberta lnfant Motor Scale (AIMS) scores were as follows: Prone position: 21; supine position: 9; sitting position: 12; standing position: 9; total score: 51; AIMS percentile: < 5; equivalent age: 11.5 mo, indicating that motor development was obviously delayed. Electromyographic evoked potential: Visual evoked potentials (VEP): The latency of bilateral P100 was normal, and brainstem auditory evoked potentials (BAEP): The peak latencies of bilateral I, III and V waves and the interpeak latencies of I-III waves and III-V waves were normal.
| Age | Gross motor | Fine motor | Adaptive ability | Language | Personal social | Comprehensive developmental quotient | Result |
| 6.9 mo | 80.0 | 80.0 | 87.0 | 72.0 | 80.0 | 80.0 | Low developmental quotient |
| 12.1 mo | 74.0 | 62.0 | 66.0 | 66.0 | 74.0 | 69.0 | Low developmental quotient |
| 14.5 mo | 72.0 | 62.0 | 62.0 | 41.0 | 62.0 | 60.0 | Low developmental quotient |
| 18.5 mo | 60.8 | 63.3 | 75.0 | 37.8 | 64.1 | 60.2 | Mild defect |
Further examinations, including 25-hydroxyvitamin D level was 54.4 ng/mL (normal range: ≥ 20 ng/mL) at the age of 6.9 mo; thyroid function: T3: 2.590 nmol/L (normal range: 1.32-4.07 nmol/L), T4: 146.200 nmol/L (normal range: 73-206 nmol/L), TSH: 5.530 mIU/L (normal range: 0.73-8.35 mIU/L), were in the normal range for the age of 8.7 mo. At 14.5 mo, neuromuscular function showed that the activity of the surface electromyographic signal (sEMG) of the gastrocnemius and adductor muscle was normal. Sensory function tests showed sensory processing disorder, chromosome karyotype analysis showed no obvious abnormalities, and blood metabolic screening (blood amino acids and acyl carnitine) was normal. Urinary metabolic screening (urine organic acid analysis) showed that 2-hydroxyisobutyric acid-2 was 1.5 (normal range: 0.0-0.5), oxalic acid-2 was 9.3 (normal range: 0.0-1.0), phosphate-3 was 132.3 (normal range: 0.0-72.7), palmitic acid-1 was 60.8 (normal range: 0.0-23.3), illustrates that insufficient energy production in the body. Otoacoustic emission (OAE) showed that both the left and right ear passed; acoustic impedance testing: both left and right ear were As, explains the pressure in the middle ear cavity is normal, the peak value decreases < 0.33 cc. Serum vitamin A level was 0.34 mg/L (normal range: 0.30-0.70 mg/L) and serum vitamin E was 11.9 mg/L (normal range: 5.0-20.0 mg/L).
At the age of 14 mo, cardiac color ultrasound showed that there was no obvious abnormality of intracardiac structure, and a normal range of left ventricular systolic function was observed. At 18.5 mo old, color ultrasound of the urinary system showed that there were no obvious abnormalities in the kidneys, ureter and bladder. At the age of 12.1 mo, digital radiography showed synpolydactyly of the right foot. At the age of 14.5 mo, magnetic resonance imaging (MRI) of the brain showed that the bilateral frontotemporal subarachnoid space was slightly wider.
A retrospective case study of the boy diagnosed with NECRC was performed in August 2022. He underwent a neurological examination, GDS, AIMS, VEP, BAEP, brain MRI, sEMG, OAE, blood and urinary metabolic screening. His intellectual disability was estimated according to the criteria of the Diagnostic and Statistical Manual of Mental Disorders-5. The clinical data of family members were obtained, investigated and independently reviewed by two neurologists.
Genomic DNA was extracted from ethylene diamine tetraacetic acid-treated peripheral blood with informed consent from the patient’s family using the QIAamp R Blood Mini Kit (Qiagen, Hilden, Germany).
The extracted DNA sample was subjected to 0.8% agarose gel electrophoresis analysis, and it was confirmed that the genomic DNA sample was free of serious degradation and impurities, and was evaluated by NanoDrop2000 and Qubit3.0 to determine the concentration and purity of DNA. The DNA was sheared with an M220 Focused-ultrasonicator (Covaris, Woburn, MA, United States), the genomic DNA was fragmented to lengths ranging from 150 to 300 bp, with an average length of 250 bp. The purified product was screened by AMPureXP magnetic beads, and the length of the screened fragment was 400 bp. The DNA target region was captured by hybridizing the genomic DNA sample library with the XGen® Exome Research Panel kit (IDT, United States), which can specifically enrich the exon region of the genome for further processing. The amplified product was purified, the library was quantified by Qubit and inspected with the Bioanalyzer2100 to detect fragment distribution of the library for sequencing. The captured and amplified DNA sample was sequenced using Illumina NovaSeq6000 (Illumina, San Diego, CA, United States) with 150 base-paired end reads.
Sequencing data were analyzed to identify disease-associated SNVs/Indels according to an in-house pipeline. Both public software and commercial packages were used during bioinformatics analysis. Raw data were processed with FASTP with adapters removing low-quality reads. The paired-end reads were then performed against the Ensemble GRCh37/hg19 reference genome with the Burrows-Wheeler Aligner. Base quality score recalibration and SNVs/Indels were conducted by the HaplotypeCaller tool of GATK after the necessary post-processes on primary alignment. SNVs/Indels were screened according to sequence depth and variant quality, and high quality and reliable variants were obtained. Notably, the online system was independently used to annotate database-based minor allele frequencies (MAFs), and the American College of Medical Genetics and Genomics (ACMG) practice guideline-based pathogenicity of every yielded gene variant was determined for conservative analysis and protein product structure prediction. Each variant was compared against several public databases, dbSNP, gnomAD, 1000 genomes project, Exome Aggregation Consortium (ExAC), Chigene in-house MAFs database and NHLBI Exome Sequencing Project 6500 (ESP6500) to achieve allele frequency in the general population. Mutationtaster, Provean, Sift, M-Cap, Polypen2_hdiv, Polypen2_hvar, and Revel software packages were used to predict protein product structure variation.
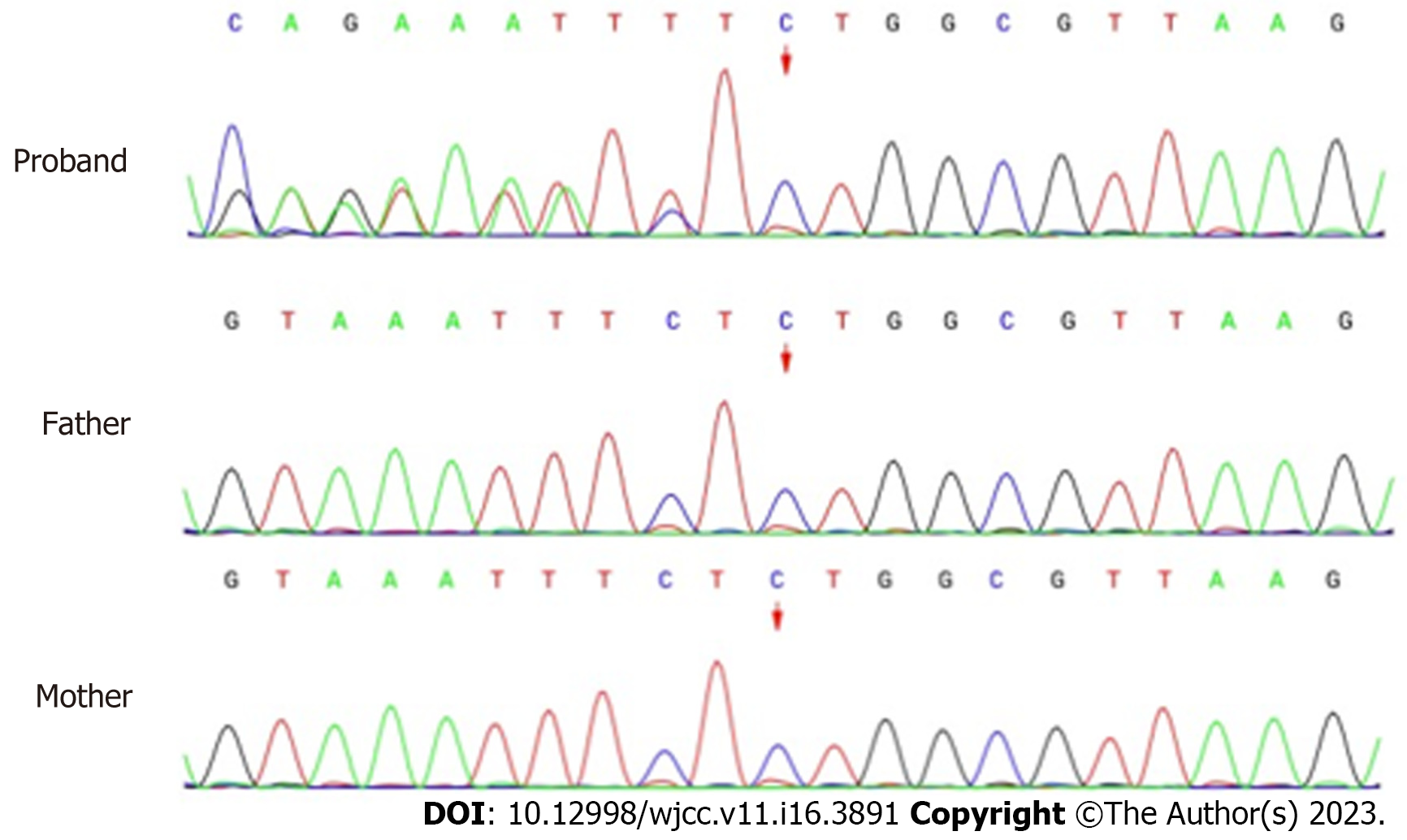
Whole-exon sequencing was performed to identify disease-causing variants. A novel heterozygous variant (NM_197968.3: c.2090_2091del, p.Ser697TrpfsTer3) of ZMYM2 gene that occurs in exon 11 was identified, as evidenced by the deletion of nucleotides from 2090 to 2091 of the ZMYM2 gene (c.2090_2091del), resulting in the change of serine at position 697 to tryptophan and downstream amino acid at position 3 was the stop codon (p.Ser697TrpfsTer3). From the pedigree of the family, it was found that neither of the parents carried the variant, they were wild-type and had no obvious clinical phenotype (Figure 2). The clinical phenotype associated with ZMYM2 gene mutation is NECRC, which has not been reported in the control population (gnomAD and ClinVar).
This frameshift variant was detected according to ACMG from the following evidence: (1) There was a frameshift variant in the ZMYM2 gene where loss of function (LOF) is a known mechanism of the disease, PVS1; (2) All sequence variants in the proband and parental samples were confirmed by Sanger sequencing analysis and the variant was de novo, and the phenotype was in accordance with the ZMYM2 gene, PS2_Moderate; and (3) The variant was absent in the control population in the Exome Sequencing Project, gnomAD, 1000 Genomes Project or ExAC, PM2_Supporting. With the evidence of PVS1 + PS2 + PM2, the class of this variant was categorized as pathogenetic.
The patient was diagnosed with NECRC and ZMYM2 gene (c.2090_2091del, p.Ser697TrpfsTer3) mutation.
Early comprehensive rehabilitation training was conducted on the basis that routine treatment might not improve the long-term outcome of NECRC patients.
Detailed history-taking and follow-up examinations 6 mo later showed that the patient was lagging behind in motor and language development.
The data in our study indicated that ZMYM2 mutation was the cause of NECRC, and the mutation site of c.2090_2091del (p.Ser697TrpfsTer3) in the ZMYM2 gene was a novel frameshift mutation. This is the first report of the ZMYM2 (c.2090_2091del) mutation in a pediatric patient and expands the genotype and phenotypic spectrum of NECRC.
ZMYM2, also known as ZNF198, FIM or RAMP, is a member of the family of MYM-type zinc finger proteins. ZMYM2 is a cellular transcription factor with a zinc finger structure coding gene which localizes to the nucleus, specifically the promyelocytic leukemia (PML) body, and is a novel B-MYB binding protein that contains 1377 amino acids with a molecular mass of 150 kDa. ZMYM2 is encoded by the zinc-finger protein that harbors 2 putative nuclear localization signals (NLS) and 10 MYM type zinc fingers, and the zinc finger domain acts as a transcription factor that mainly binds to the targets DNA and RNA. The zinc finger structure also mediates protein-protein interactions to regulate the efficiency of binding nucleic acids[1]. The N-terminal of ZMYM2 includes 3 action sites related to Small Ubiquitin Like Modifier, including a MYM zinc finger domain and a proline/valine-rich domain, is related to the formation and stabilization of PML nuclear bodies (PML-NBs), the C-terminal acidic domain contains a putative NLS and a region similar to Cre-likedomain[1-4], and mutant mRNA transcripts are predicted to undergo nonsense-mediated decay, which prevents its translation, leading to ZMYM2 haploinsufficiency and LOF[5].
ZMYM2 has a specialized role in pronephric development in a subset of regions, LOF variants in ZMYM2 induce CAKUT-like defects. Quantification of sites with morpholino oli
We also reviewed the literature[5] and found 15 different heterozygous nonsense or frameshift mutations of ZMYM2 in 16 unrelated families, and 19 affected individuals with CAKUT and/or syndromic extra-renal features, and the heterozygous truncated mutations affected reproductive function. The genes related to the single gene form of CAKUT accounted for 14%-20% of cases[6-8], neurological manifestations were noted in 17 affected individuals in 15 unrelated families, including 5 individuals with intellectual disability, 10 individuals with motor retardation, 5 individuals with speech delay, 8 individuals with urinary system abnormalities, 6 individuals with heart abnormalities, 4 individuals with hypodystonia and 5 individuals with microcephaly.
ZMYM2 selectively binds to the LSD1–CoREST– HDAC1 ternary complex, which is characterized as a corepressor of transcription by interacting with different nuclear receptors, and is associated with LSD1-containing corepressor complexes, such as the LSD1-CoREST-HDAC1 complex on chromatin to regulate gene expression[5,9]. There are multiple ZMYM2 interactors, including members of the LSD1- CoREST-HDAC1 pathway, suggesting that the wider range of ZMYM2 interaction groups, including DNA-binding transcription factors, transcriptional-corepressors, and proteins related to chromatin regulation and tissue, represent potential candidates for urinary tract malformations[3].
FOXP transcription factors play important roles in neurodevelopment, and FOXP cooperatively regulates gene expression by forming homo- and hetero-dimers with each other. ZMYM2 is a novel FOXP-interacting transcription factor[10], and other genes in this interaction group can also be considered candidates for participation in the disease due to FOXP1 or ZMYM2 LOF mutations. Most pathogenic pathways of ZMYM2-like proteins remain elusive, and further work is required to clarify the role of potential interactions in the pathogenesis of NECRC caused by ZMYM2 mutations.
Relatively few genetic findings have reached the clinic, and in the wide array of inherited metabolic disorders, NECRC has attracted increasing attention due to the neurological damage it causes. The ZMYM2 gene (c.2090_2091del, p.Ser697TrpfsTer3) mutation induces a variety of clinical phenotypes, such as neurodevelopmental disorders, intellectual disability, autism spectrum disorder, schizophrenia, congenital heart defects, hydroureter, duplex and cystic kidneys. For patients with suspected NECRC, accurate molecular diagnosis can be provided promptly in children carrying the genetic mutation, which is of great significance for early intervention, precise treatment and family genetic counseling on NECRC.
The authors are thankful to the patient and his family for making this study possible. Responsibility for interpretation of data, conclusions and opinions lies with the author.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Medicine, research and experimental
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Velikova TV, Bulgaria; Watanabe T, Japan S-Editor: Liu XF L-Editor: Webster JR P-Editor: Zhang YL
| 1. | Mackay JP, Crossley M. Zinc fingers are sticking together. Trends Biochem Sci. 1998;23:1-4. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 338] [Cited by in F6Publishing: 370] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 2. | Still IH, Cowell JK. The t(8;13) atypical myeloproliferative disorder: further analysis of the ZNF198 gene and lack of evidence for multiple genes disrupted on chromosome 13. Blood. 1998;92:1456-1458. [PubMed] [Cited in This Article: ] |
| 3. | Gocke CB, Yu H. ZNF198 stabilizes the LSD1-CoREST-HDAC1 complex on chromatin through its MYM-type zinc fingers. PLoS One. 2008;3:e3255. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 61] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 4. | Kunapuli P, Kasyapa CS, Chin SF, Caldas C, Cowell JK. ZNF198, a zinc finger protein rearranged in myeloproliferative disease, localizes to the PML nuclear bodies and interacts with SUMO-1 and PML. Exp Cell Res. 2006;312:3739-3751. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 26] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 5. | Connaughton DM, Dai R, Owen DJ, Marquez J, Mann N, Graham-Paquin AL, Nakayama M, Coyaud E, Laurent EMN, St-Germain JR, Blok LS, Vino A, Klämbt V, Deutsch K, Wu CW, Kolvenbach CM, Kause F, Ottlewski I, Schneider R, Kitzler TM, Majmundar AJ, Buerger F, Onuchic-Whitford AC, Youying M, Kolb A, Salmanullah D, Chen E, van der Ven AT, Rao J, Ityel H, Seltzsam S, Rieke JM, Chen J, Vivante A, Hwang DY, Kohl S, Dworschak GC, Hermle T, Alders M, Bartolomaeus T, Bauer SB, Baum MA, Brilstra EH, Challman TD, Zyskind J, Costin CE, Dipple KM, Duijkers FA, Ferguson M, Fitzpatrick DR, Fick R, Glass IA, Hulick PJ, Kline AD, Krey I, Kumar S, Lu W, Marco EJ, Wentzensen IM, Mefford HC, Platzer K, Povolotskaya IS, Savatt JM, Shcherbakova NV, Senguttuvan P, Squire AE, Stein DR, Thiffault I, Voinova VY, Somers MJG, Ferguson MA, Traum AZ, Daouk GH, Daga A, Rodig NM, Terhal PA, van Binsbergen E, Eid LA, Tasic V, Rasouly HM, Lim TY, Ahram DF, Gharavi AG, Reutter HM, Rehm HL, MacArthur DG, Lek M, Laricchia KM, Lifton RP, Xu H, Mane SM, Sanna-Cherchi S, Sharrocks AD, Raught B, Fisher SE, Bouchard M, Khokha MK, Shril S, Hildebrandt F. Mutations of the Transcriptional Corepressor ZMYM2 Cause Syndromic Urinary Tract Malformations. Am J Hum Genet. 2020;107:727-742. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 6. | van der Ven AT, Connaughton DM, Ityel H, Mann N, Nakayama M, Chen J, Vivante A, Hwang DY, Schulz J, Braun DA, Schmidt JM, Schapiro D, Schneider R, Warejko JK, Daga A, Majmundar AJ, Tan W, Jobst-Schwan T, Hermle T, Widmeier E, Ashraf S, Amar A, Hoogstraaten CA, Hugo H, Kitzler TM, Kause F, Kolvenbach CM, Dai R, Spaneas L, Amann K, Stein DR, Baum MA, Somers MJG, Rodig NM, Ferguson MA, Traum AZ, Daouk GH, Bogdanović R, Stajić N, Soliman NA, Kari JA, El Desoky S, Fathy HM, Milosevic D, Al-Saffar M, Awad HS, Eid LA, Selvin A, Senguttuvan P, Sanna-Cherchi S, Rehm HL, MacArthur DG, Lek M, Laricchia KM, Wilson MW, Mane SM, Lifton RP, Lee RS, Bauer SB, Lu W, Reutter HM, Tasic V, Shril S, Hildebrandt F. Whole-Exome Sequencing Identifies Causative Mutations in Families with Congenital Anomalies of the Kidney and Urinary Tract. J Am Soc Nephrol. 2018;29:2348-2361. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 128] [Cited by in F6Publishing: 127] [Article Influence: 21.2] [Reference Citation Analysis (0)] |
| 7. | Weber S, Moriniere V, Knüppel T, Charbit M, Dusek J, Ghiggeri GM, Jankauskiené A, Mir S, Montini G, Peco-Antic A, Wühl E, Zurowska AM, Mehls O, Antignac C, Schaefer F, Salomon R. Prevalence of mutations in renal developmental genes in children with renal hypodysplasia: results of the ESCAPE study. J Am Soc Nephrol. 2006;17:2864-2870. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 267] [Cited by in F6Publishing: 242] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 8. | Rasmussen M, Sunde L, Nielsen ML, Ramsing M, Petersen A, Hjortshøj TD, Olsen TE, Tabor A, Hertz JM, Johnsen I, Sperling L, Petersen OB, Jensen UB, Møller FG, Petersen MB, Lildballe DL. Targeted gene sequencing and whole-exome sequencing in autopsied fetuses with prenatally diagnosed kidney anomalies. Clin Genet. 2018;93:860-869. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 40] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 9. | Mojarad BA, Yin Y, Manshaei R, Backstrom I, Costain G, Heung T, Merico D, Marshall CR, Bassett AS, Yuen RKC. Genome sequencing broadens the range of contributing variants with clinical implications in schizophrenia. Transl Psychiatry. 2021;11:84. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in F6Publishing: 12] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 10. | Estruch SB, Graham SA, Quevedo M, Vino A, Dekkers DHW, Deriziotis P, Sollis E, Demmers J, Poot RA, Fisher SE. Proteomic analysis of FOXP proteins reveals interactions between cortical transcription factors associated with neurodevelopmental disorders. Hum Mol Genet. 2018;27:1212-1227. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in F6Publishing: 35] [Article Influence: 7.0] [Reference Citation Analysis (0)] |