Copyright
©The Author(s) 2022.
World J Clin Cases. Oct 6, 2022; 10(28): 10017-10030
Published online Oct 6, 2022. doi: 10.12998/wjcc.v10.i28.10017
Published online Oct 6, 2022. doi: 10.12998/wjcc.v10.i28.10017
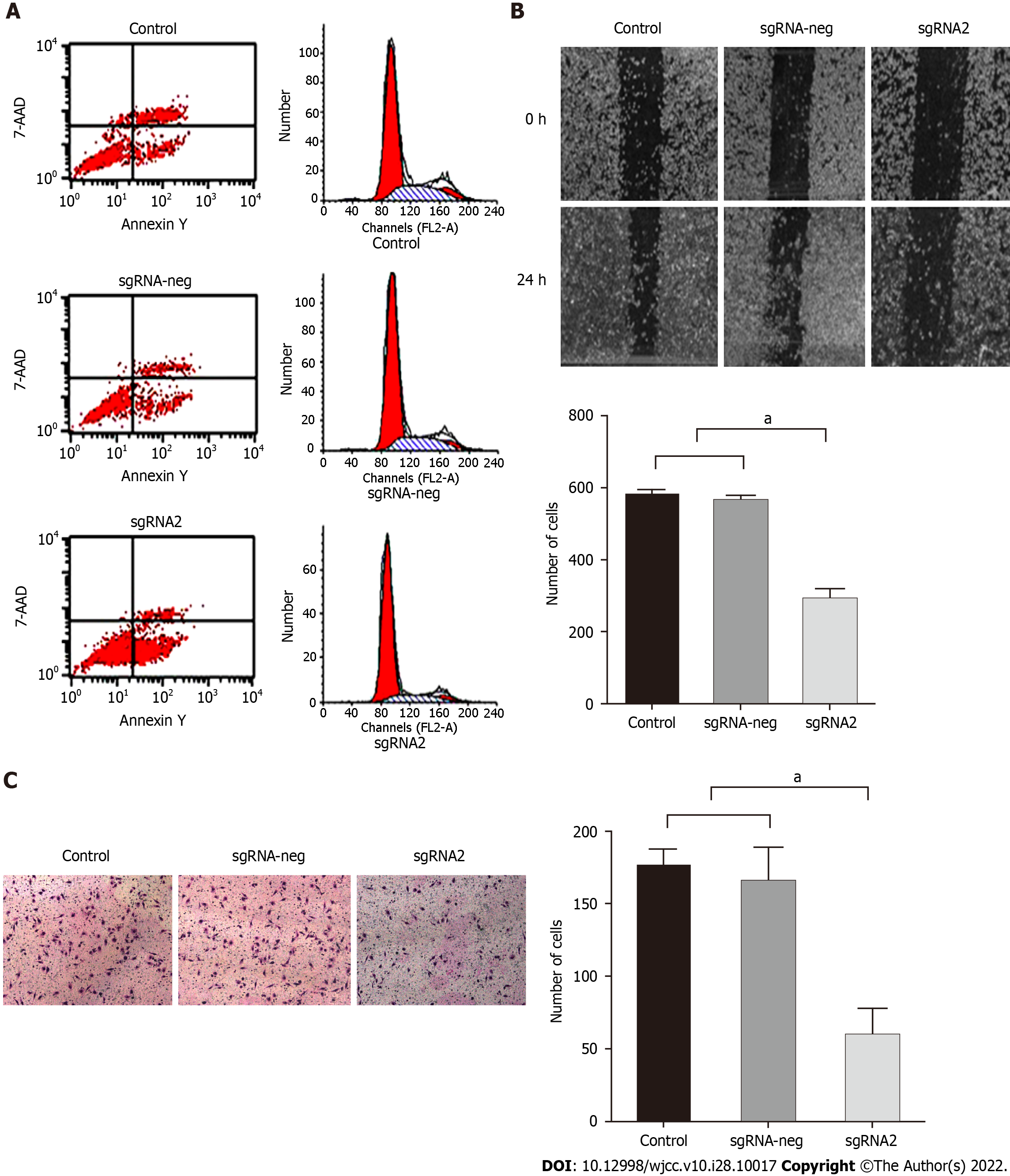
Figure 3 Editing IGF-1R on effects of biological features of HepG2 cells.
HepG2 cells were divided into control, sgRNA-neg, and sgRNA2 groups. After HepG2 cells transfected with sgRNA2 plasmids: A: Number of apoptotic cells in the sgRNA2 group was significantly higher than those in the sgRNA-neg or control group. B: Migration rates of HepG2 cells were detected by scratch wound test and suppressive effect of specific sgRNA2-mediated IGF-1R on the migration potential of HepG2 cells. Left: Representative images of migration cells at 0 h or at24 h from the control, sgRNA-neg, and sgRNA group; Right: Comparative analysis of migration cells among the different group. C: Invasion rates of HepG2 cells for 24 h were detected by Transwell assay and suppressive effect of sgRNA2-mediated IGF-1R on the invasion potential of HepG2 cells. Invaded cells were enumerated under a light microscope (magnification × 100). Left: Representative images of invasive cells stained with crystal violet from the control, sgRNA-neg, and sgRNA group; Right: Comparative analysis of invasive cells among the different group. P < 0.001 vs Control or sgRNA-neg group. The data were shown as mean ± SD. IGF-IR: Insulin-like growth factor-1 receptor. aP < 0.05.
- Citation: Yao M, Cai Y, Wu ZJ, Zhou P, Sai WL, Wang DF, Wang L, Yao DF. Effects of targeted-edited oncogenic insulin-like growth factor-1 receptor with specific-sgRNA on biological behaviors of HepG2 cells. World J Clin Cases 2022; 10(28): 10017-10030
- URL: https://www.wjgnet.com/2307-8960/full/v10/i28/10017.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v10.i28.10017