Copyright
©The Author(s) 2015.
World J Clin Cases. Jul 16, 2015; 3(7): 575-598
Published online Jul 16, 2015. doi: 10.12998/wjcc.v3.i7.575
Published online Jul 16, 2015. doi: 10.12998/wjcc.v3.i7.575
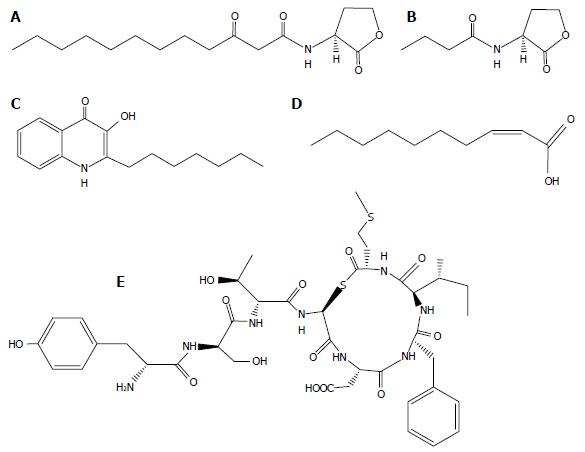
Figure 1 Structures of representative quorum sensing signal molecules of Pseudomonas aeruginosa.
A: 3-oxo-C12-homoserine lactone; B: N-butyryl-L-homoserine lactone; C: 2-heptyl-3-hydroxy-4-quinolone; D: DSF-like fatty acids, cis-2-decenoic acid) and S. aureus (E: AIP group I).
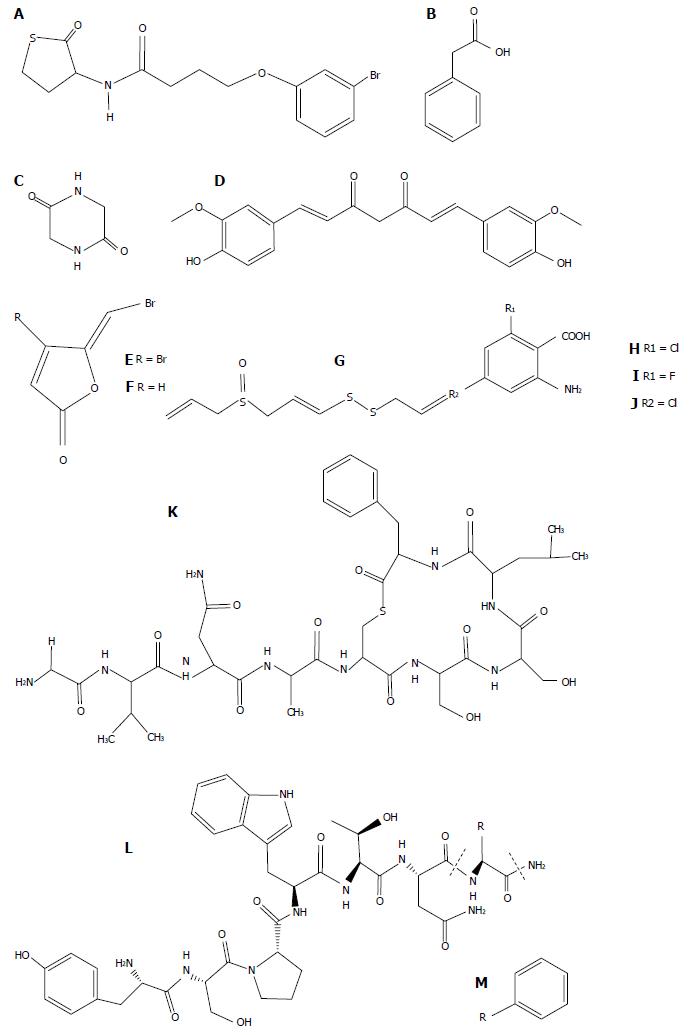
Figure 2 Structures of quorum sensing inhibitors evaluated in animal models.
A: Meta-bromo-thiolactone; B: Phenylacetic acid; C: 2,5-piperazinedione; D: Cucurmin; E: Furanone C-30; F: Furanone C-56; G: Ajoene 4,5,9,-trithiadodeca-1,6,11-triene-9-oxide; H: 2-amino-6-chlorobenzoic acid; I: 2-amino-6-fluorobenzoic acid; J: 2-amino-4-chlorobenzoic acid; K: AIP-II; L: RIP (H-Tyr-Ser-Pro-Trp-Thr-Asn-Phe-NH2); M: FS8 (H-Tyr-Ser-Pro-Trp-Thr-Asn-Ala-NH2).
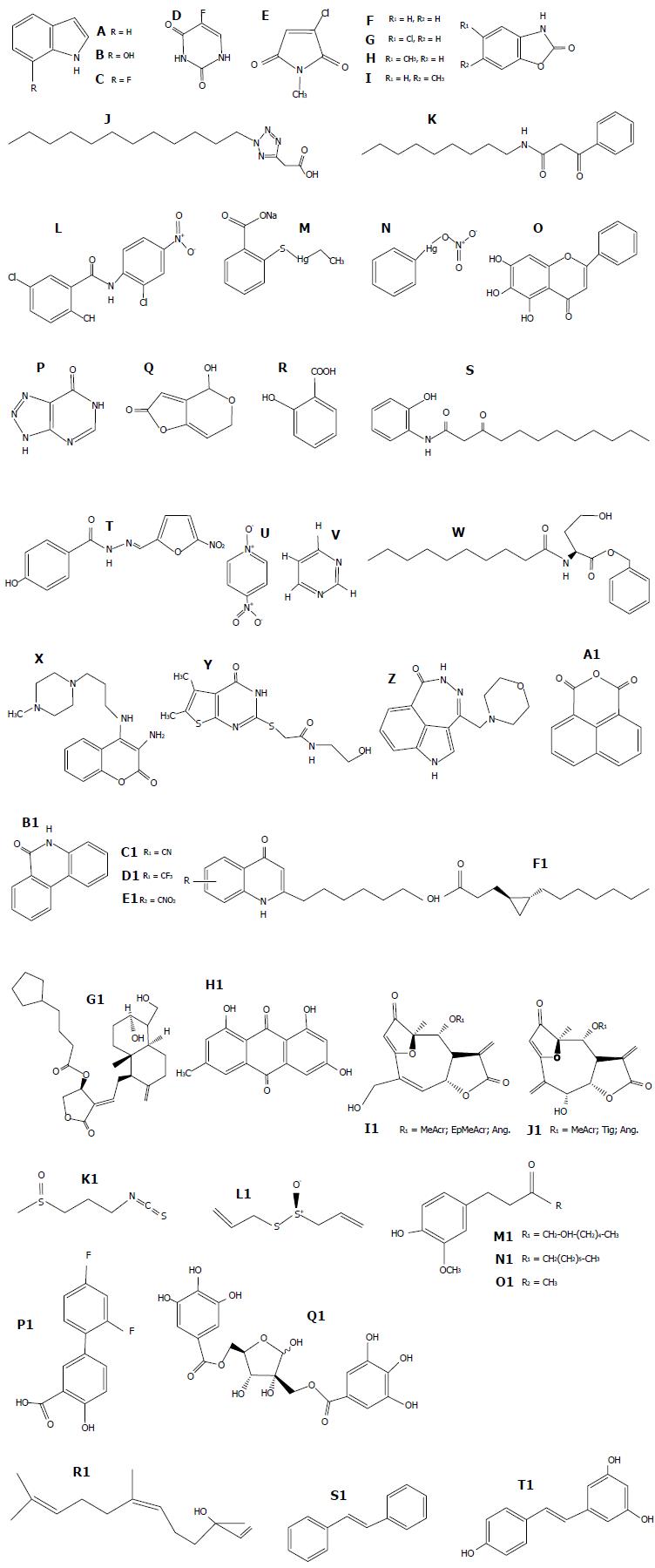
Figure 3 Structures of representative quorum quenching molecules of Pseudomonas aeruginosa.
A: Indole; B: 7-hidroxy indole; C: 7-fluoroindole; D: 5-fluorouracil; E: 2-chloro-N-methyl-maleimide; F: 1,3-benzoxazol-2(3H)-one; G: 5-cloro-1,3-benzoxazol-2(3H)-one (clorzoxazone); H: 5-methyl-1,3-benzoxazol-2(3H)-one; I: 6-methyl-1,3-benzoxazol-2(3H)-one; J: PD12; K: V-06-018; L: Niclosamide; M: Thimerosal; N: Phenylmercuric nitrate; O: Baicalein; P: 5-imino-4,6-dihydro-3H-1,2,3-triazolo[5,4-d]pyrimidin-7-one; Q: Patulin; R: Salicylic acid; S: 3-oxo-C12-(2-aminophenol); T: Nifuroxazide; U: 4-nitropyridine-N-oxide; V: Pyrimidine; W: N-decanoyl-L-homoserine benzyl ester; X: V23; Y: V30; Z: P1; A1: NAP; B1: PJ97A; C1: 6-CN; D1: 6-CF3; E1: 6-NO2; F1: Lyngbyoic acid; G1: Andrographolide 14-(5-cyclopentylvaleryl); H1: Emodin; I1: Goyazensolide-type; J1: Isogoyazensolide-type; K1: Iberin; L1: Allicin; M1: [6]-gingerol; N1: [6]-shogaol; O1: Zingerone and S. aureus; P1: Diflunisal; Q1: Hamamelitannin; R1: Cis-nerolidol; S1: Trans-stilbene; T1: Resveratrol.
- Citation: Castillo-Juárez I, Maeda T, Mandujano-Tinoco EA, Tomás M, Pérez-Eretza B, García-Contreras SJ, Wood TK, García-Contreras R. Role of quorum sensing in bacterial infections. World J Clin Cases 2015; 3(7): 575-598
- URL: https://www.wjgnet.com/2307-8960/full/v3/i7/575.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v3.i7.575