Copyright
©The Author(s) 2024.
World J Clin Cases. Mar 26, 2024; 12(9): 1622-1633
Published online Mar 26, 2024. doi: 10.12998/wjcc.v12.i9.1622
Published online Mar 26, 2024. doi: 10.12998/wjcc.v12.i9.1622
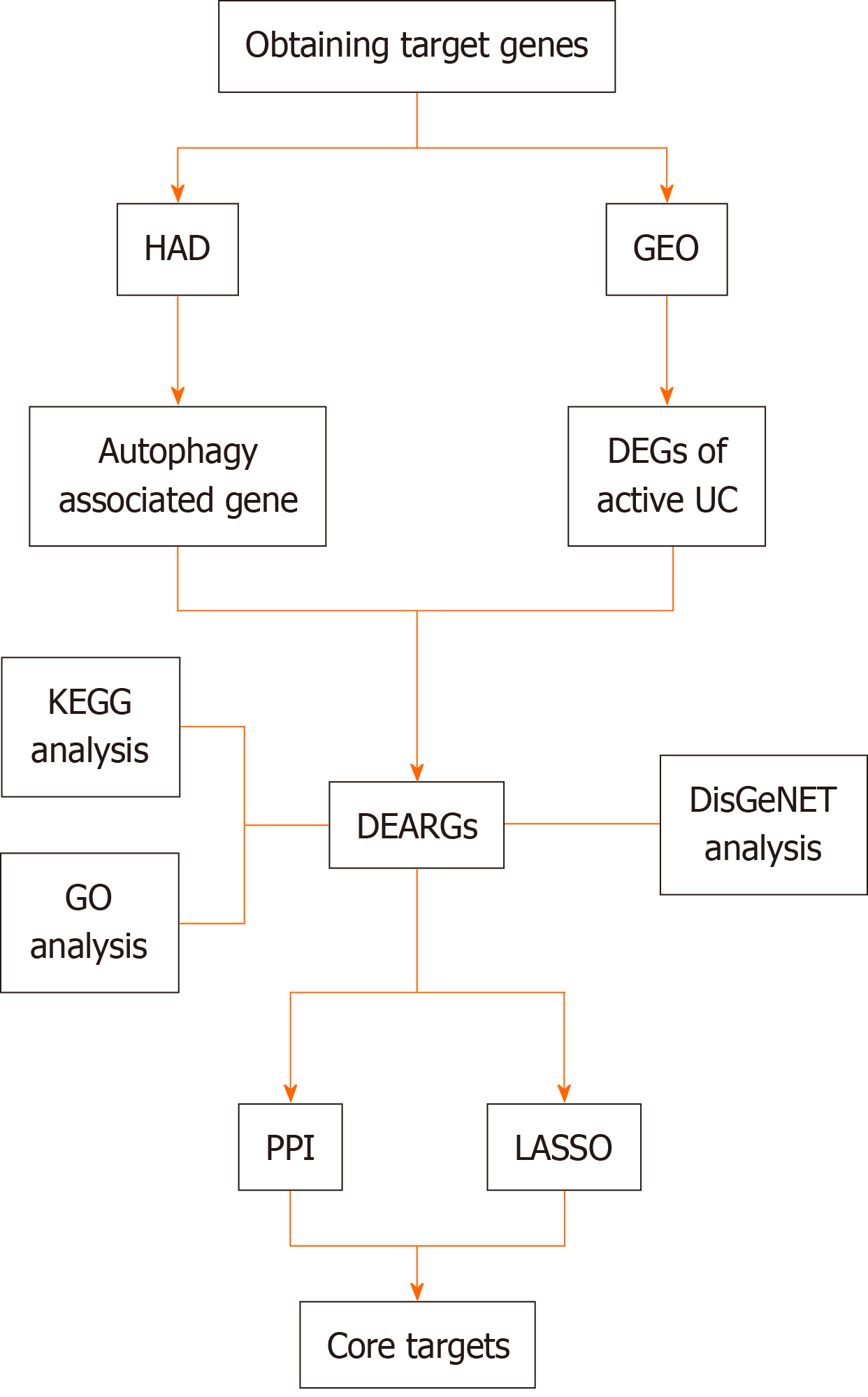
Figure 1 Study flow chart.
HAD: Human Autophagy Database; GEO: Gene Expression Omnibus database; UC: Ulcerative colitis; DEGs: Differentially expressed genes; DEARGs: Differentially expressed autophagy-related genes; KEGG: Kyoto Encyclopedia of Genes and Genomes; GO: Gene Ontology; PPI: Protein-Protein Interaction; LASSO: Least absolute shrinkage and selection operator.
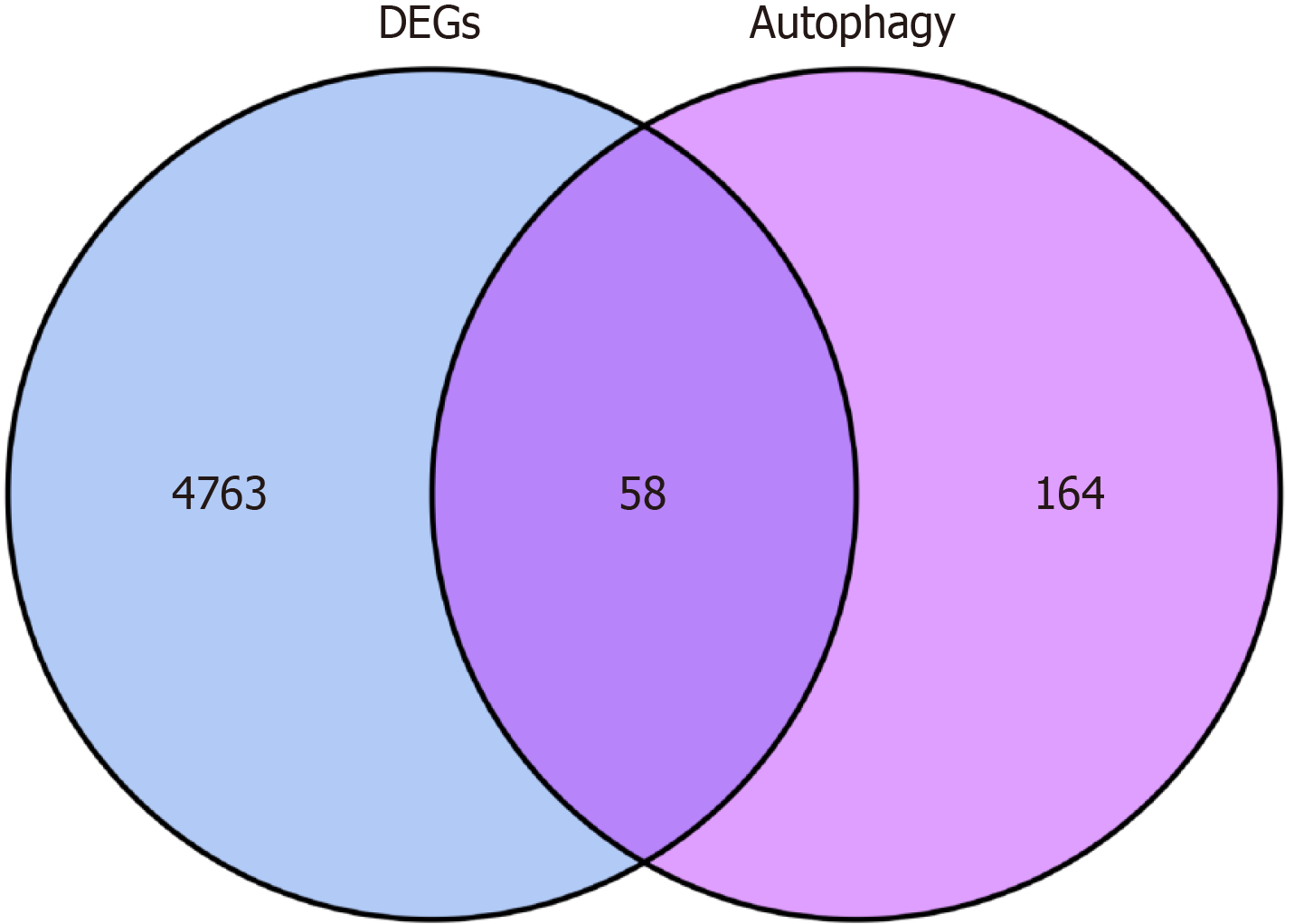
Figure 2 Intersection of autophagy-related targets and differentially expressed genes.
There are 4821 differentially expressed genes and 232 autophagy-related targets. A total of 58 differentially expressed autophagy-related genes intersected. DEGs: Differentially expressed genes.
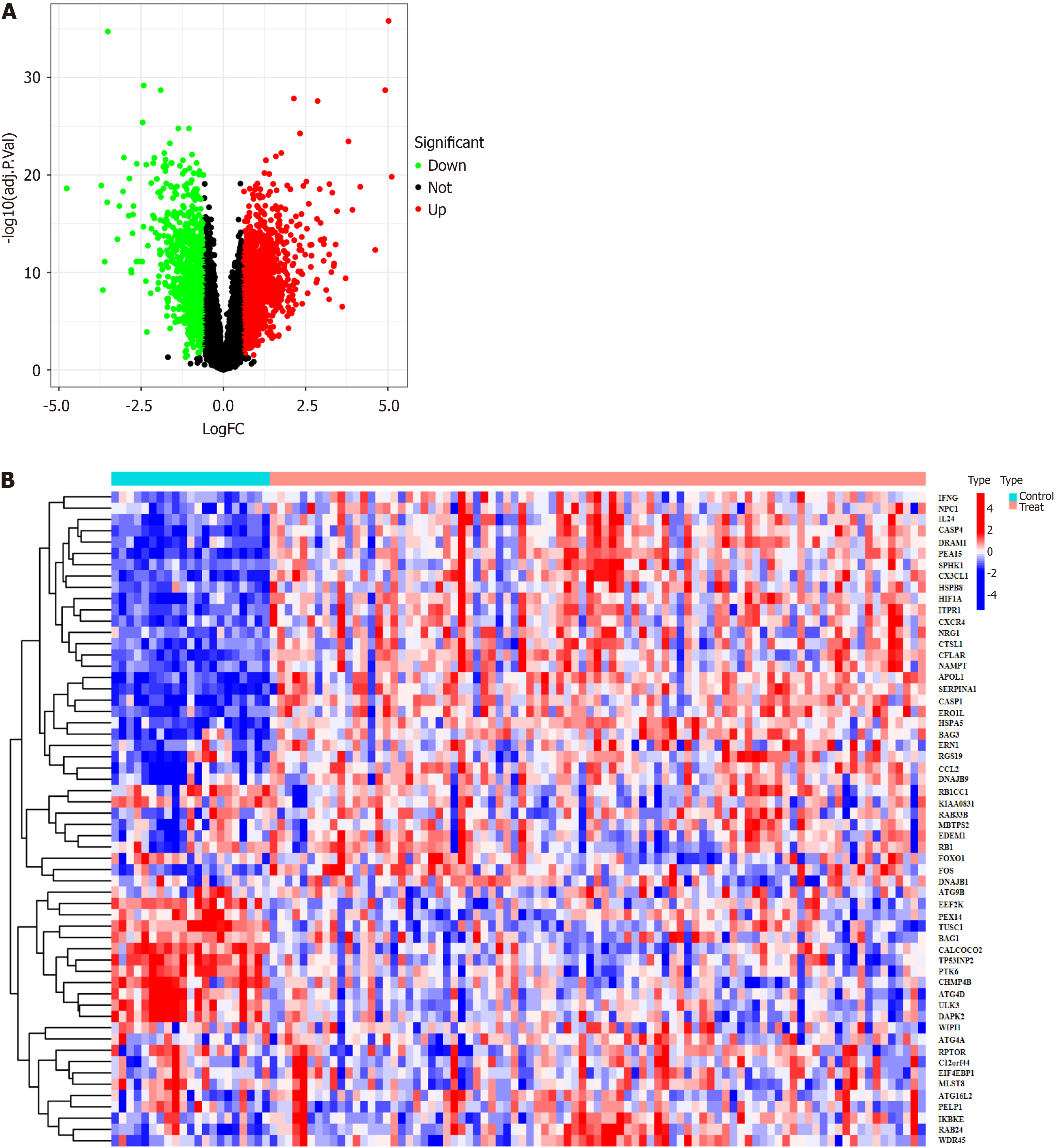
Figure 3 Volcano map and heat map showing expression of the differentially expressed autophagy-related genes.
A: The downregulated targets are represented by green dots, the upregulated targets are represented by red dots, and the black dots indicate no significant difference in expression between active ulcerative colitis patients and healthy controls. Heat map showing expression of the differentially expressed autophagy-related genes; B: The blue color indicates low expression, while the red color indicates high expression.
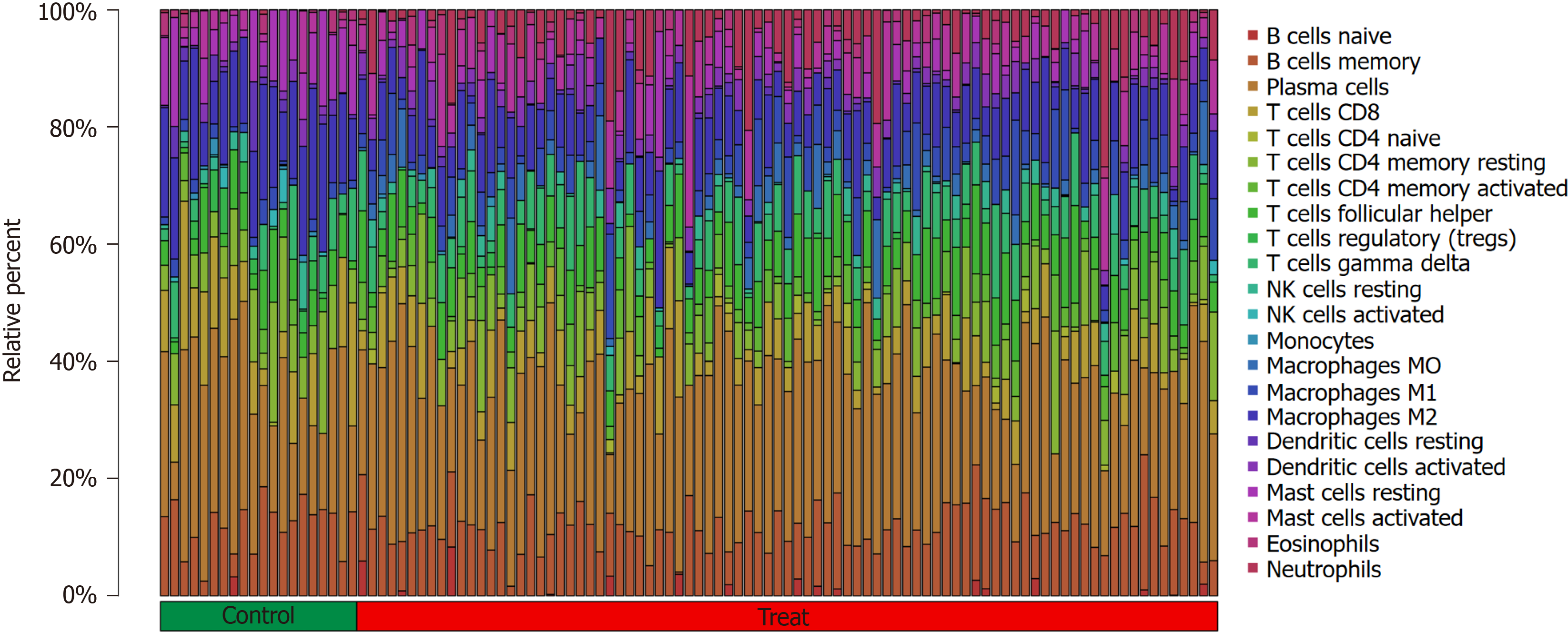
Figure 4 Proportion of immune cells in samples from the active ulcerative colitis group and healthy controls.
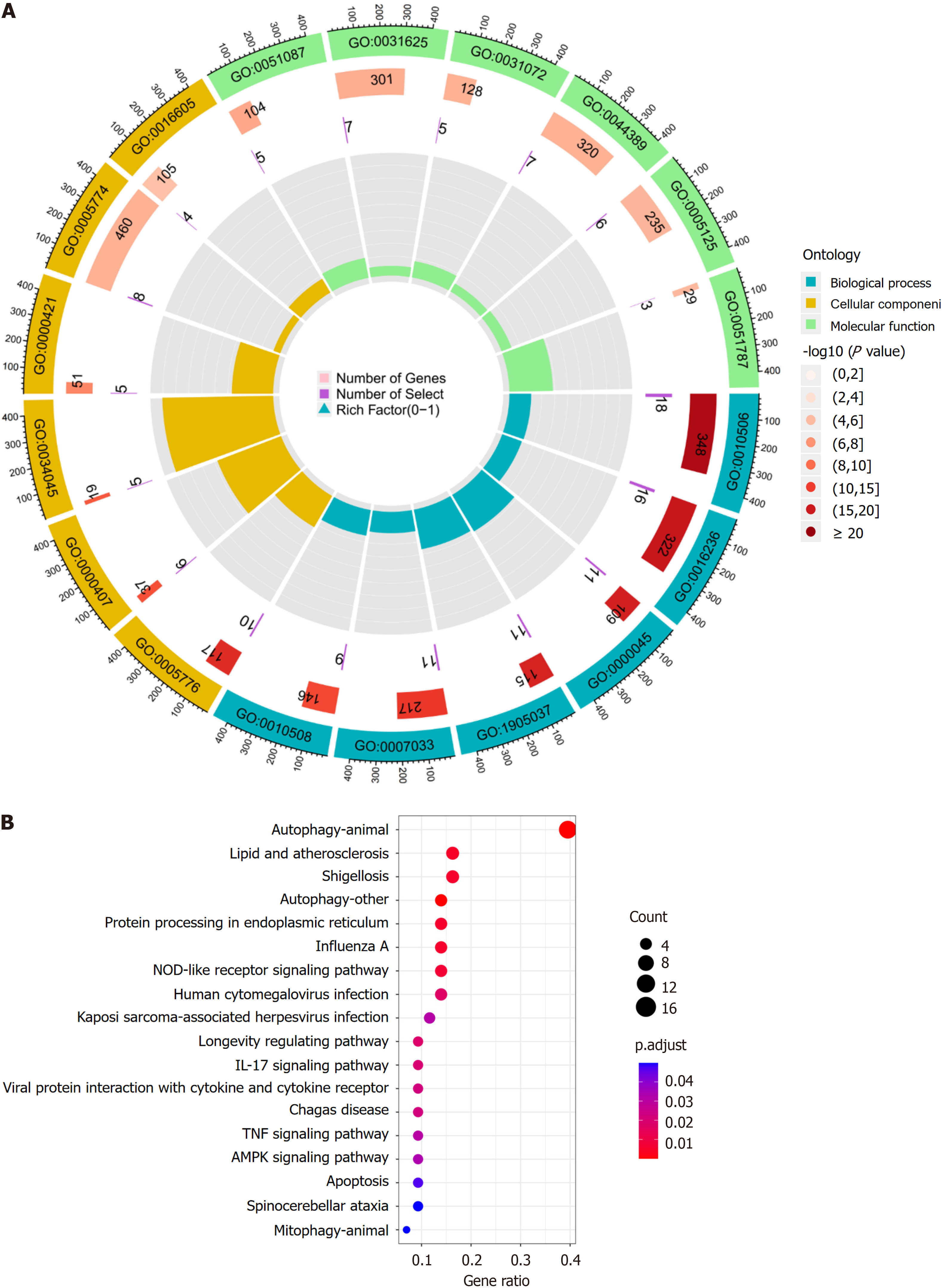
Figure 5 Results of gene ontology enrichment analysis.
A: The blue part represents biological processes, the yellow part represents cellular components and the green part represents molecular functions. the height of the bar in the inner circle is proportional to the degree of enrichment. Results of Kyoto Encyclopedia of Genes and Genomes enrichment analysis; B: The horizontal coordinate is the number of enriched differentially expressed autophagy-related genes and the color of the node changes from red to purple according to the adjustment.
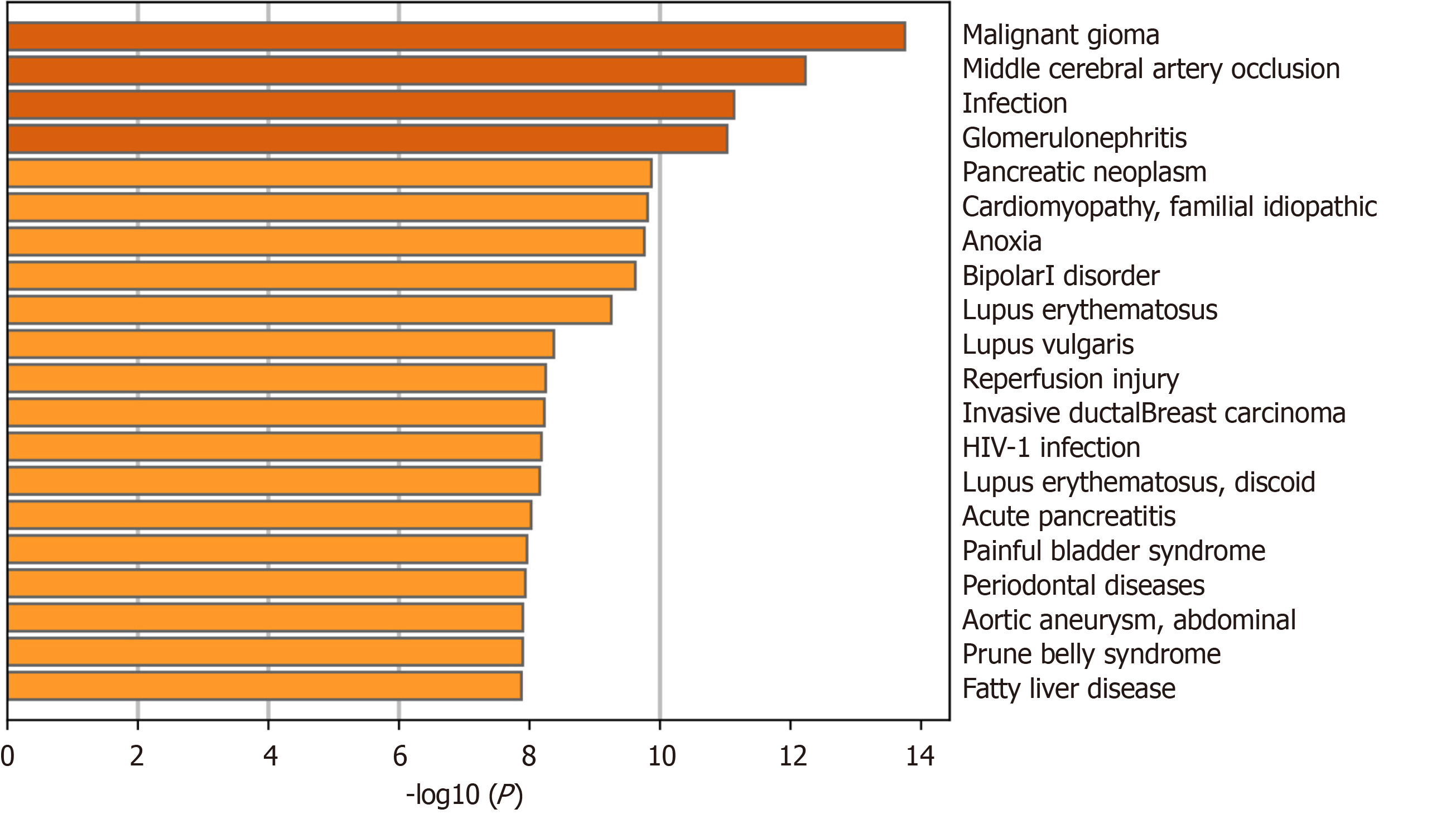
Figure 6 Validation of core targets in DisGeNET.
The more yellow the color of the bar, the higher the correlation.
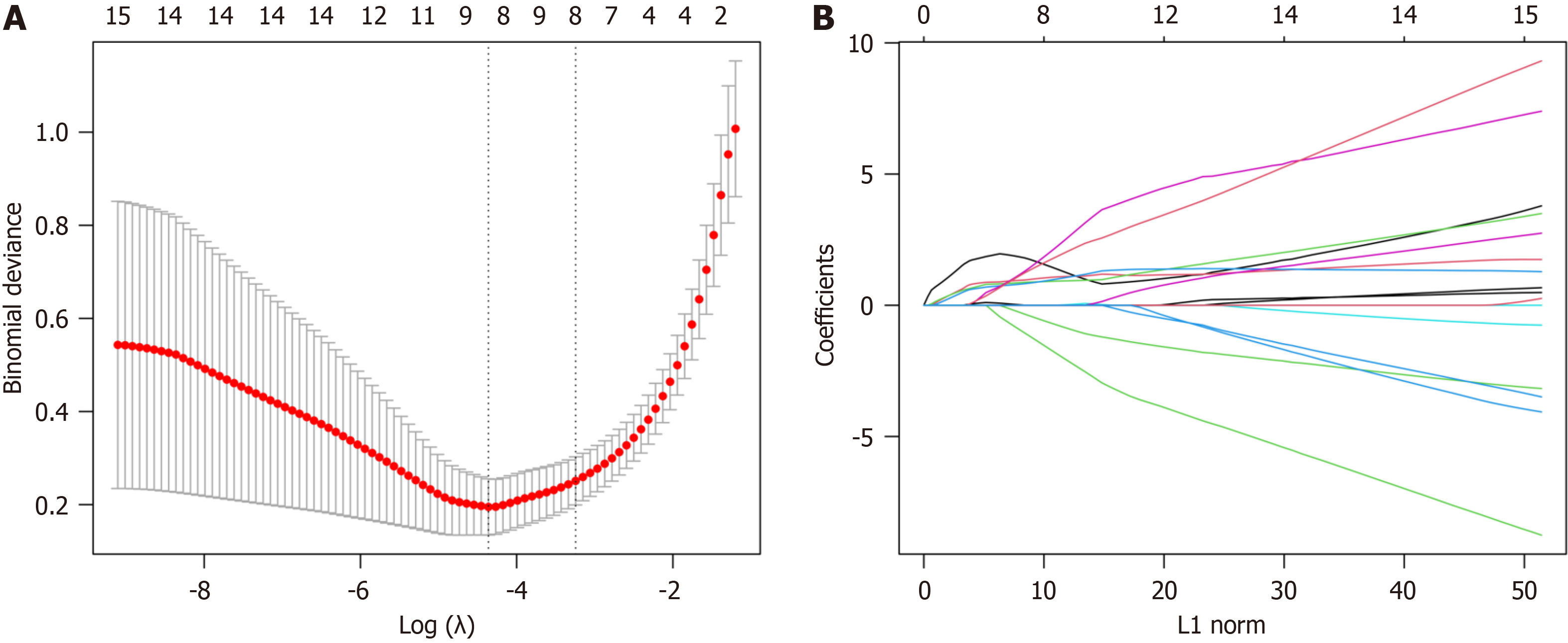
Figure 7 Results of least absolute shrinkage and selection operator regression model.
A: Cross-validation error curve; B: Least absolute shrinkage and selection operator regression coefficient path plot.
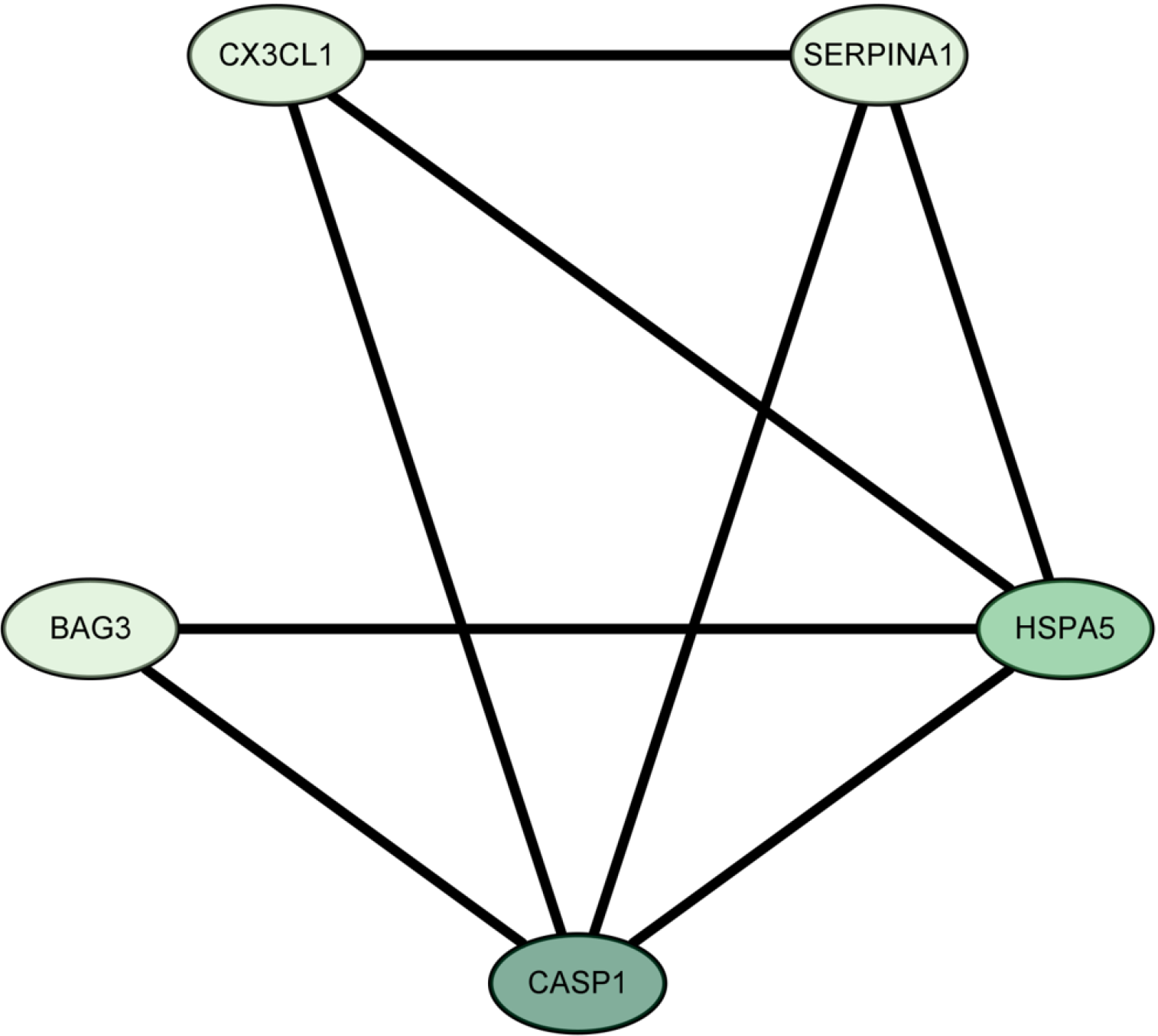
Figure 8 The five core targets and their relationship.
The greener the color of the node, the higher the value of the degree, and the more important the relationship.
- Citation: Gong ZZ, Li T, Yan H, Xu MH, Lian Y, Yang YX, Wei W, Liu T. Exploring the autophagy-related pathogenesis of active ulcerative colitis. World J Clin Cases 2024; 12(9): 1622-1633
- URL: https://www.wjgnet.com/2307-8960/full/v12/i9/1622.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v12.i9.1622