Copyright
©The Author(s) 2022.
World J Clin Cases. May 16, 2022; 10(14): 4446-4459
Published online May 16, 2022. doi: 10.12998/wjcc.v10.i14.4446
Published online May 16, 2022. doi: 10.12998/wjcc.v10.i14.4446
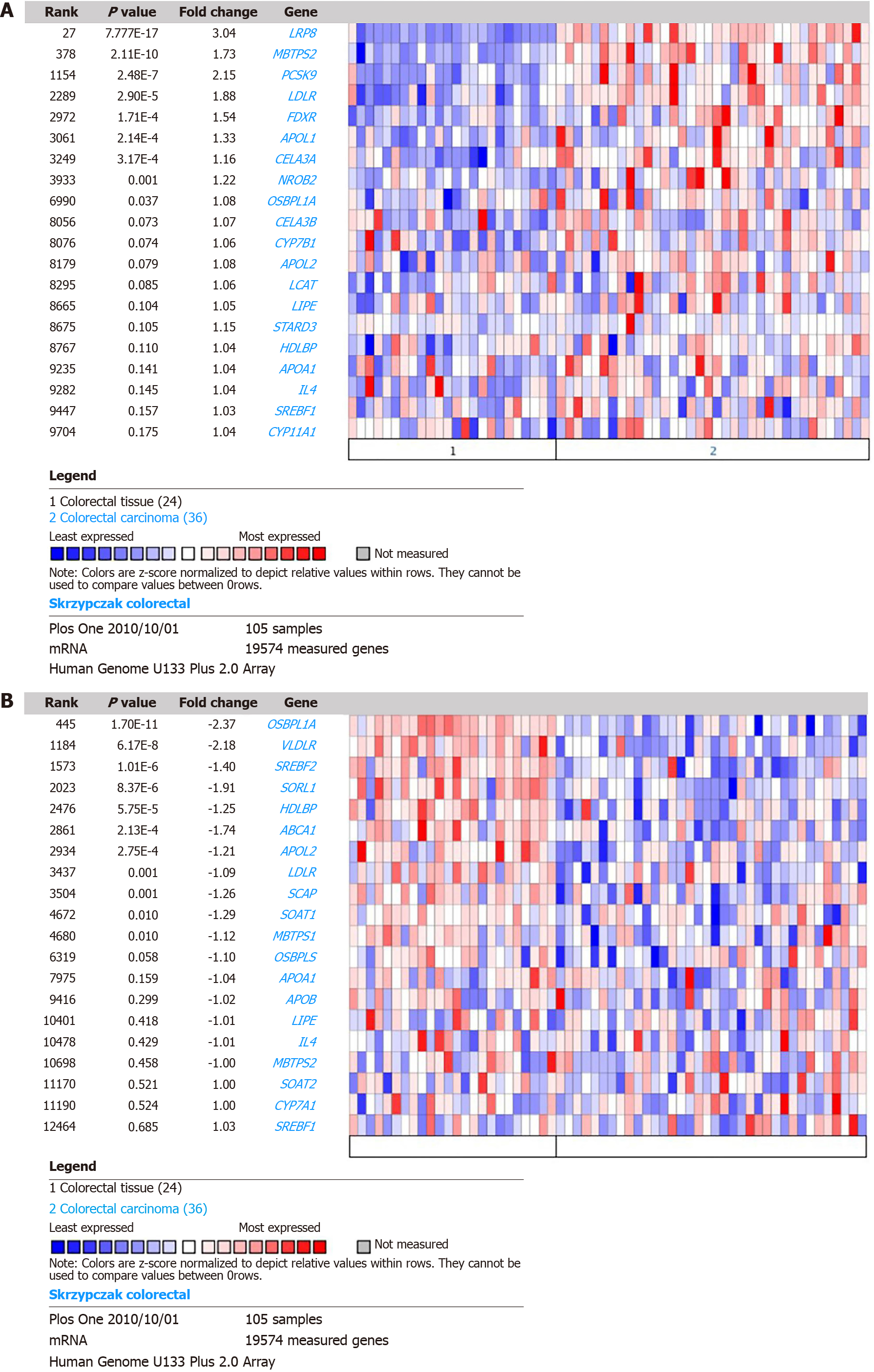
Figure 1 Comparison of concept: “Cholesterol metabolism - go biological process” in skrzypczak colorectal.
A: Upregulated expression of genes in the cholesterol metabolism pathway of colorectal cancer (CRC); B: Downregulated expression of genes in the cholesterol metabolism pathway of CRC.
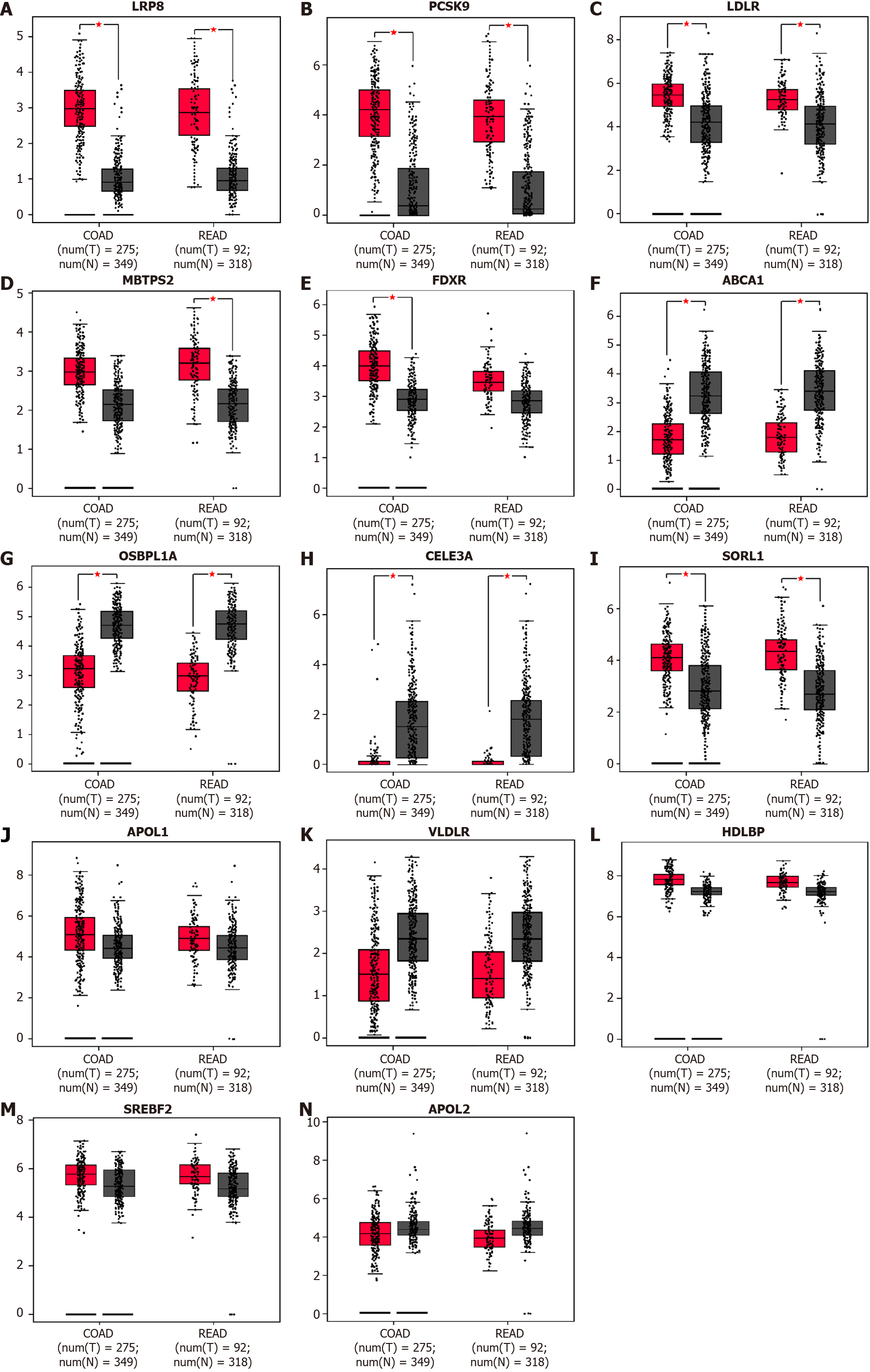
Figure 2 Identification of candidate differentially expressed genes.
The mRNA expression of differentially expressed genes between tumor and normal tissues was plotted by Gene Expression Profiling Interactive Analysis (GEPIA) with data from the The Cancer Genome Atlas database. A-C: Lipoprotein receptor-related protein 8, PCSK9, and low-density lipoprotein receptor (LDLR) were upregulated in colorectal cancer tissue compared with normal tissue; D, E: MBTPS2 and FDXR showed significantly higher expression only in rectal cancer and colon cancer, respectively; F, G: ABCA1 and OSBPL1A were downregulated in CRC tissue, consistent with the results from Oncomine; H: CELA3A was downregulated; I: SORL1 was upregulated in cancer tissue, which is the reverse of the above results; J-N: The expression levels of APOL1, VLDLR, HDLBP, SREBF2 and APOL2 were comparable between cancer and normal tissues according to the GEPIA analysis results.
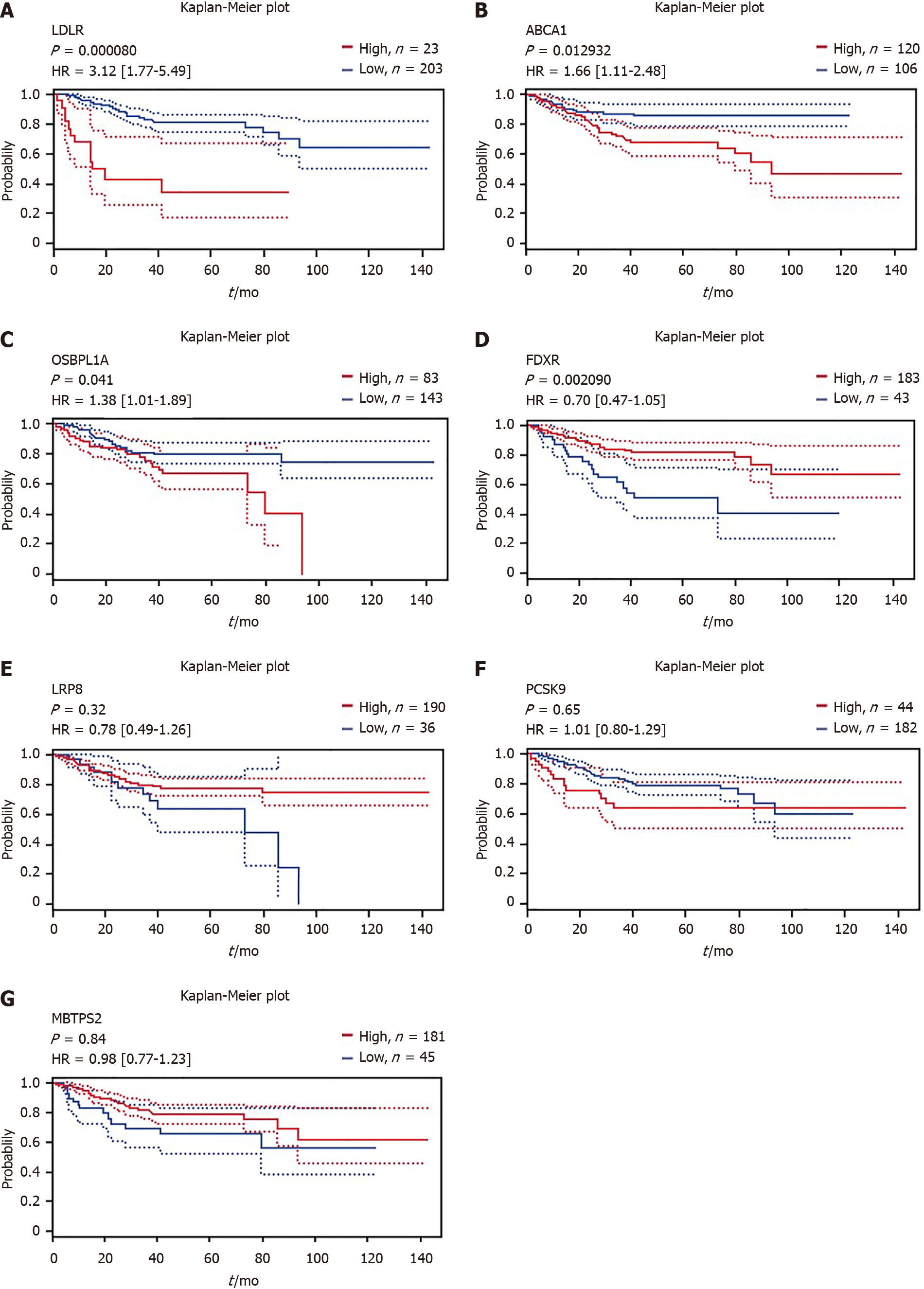
Figure 3 Correlation of differentially expressed gene expression and disease-free survival in patients with colorectal cancer.
The survival curves comparing the patients with high (red) and low (blue) expression were plotted from the PrognoScan database. A-C: High mRNA expression of low-density lipoprotein receptor, ABCA1 and OSBPL1A was an unfavorable prognostic factor for disease-free survival (DFS) in colorectal cancer (CRC) patients; D: High mRNA expression of FDXR was a favorable prognostic factor for DFS in CRC patients; E-G: Lipoprotein receptor-related protein 8, PCSK9 and MBTPS2 expression could not be used to predict DFS outcome according to the results of this analysis.
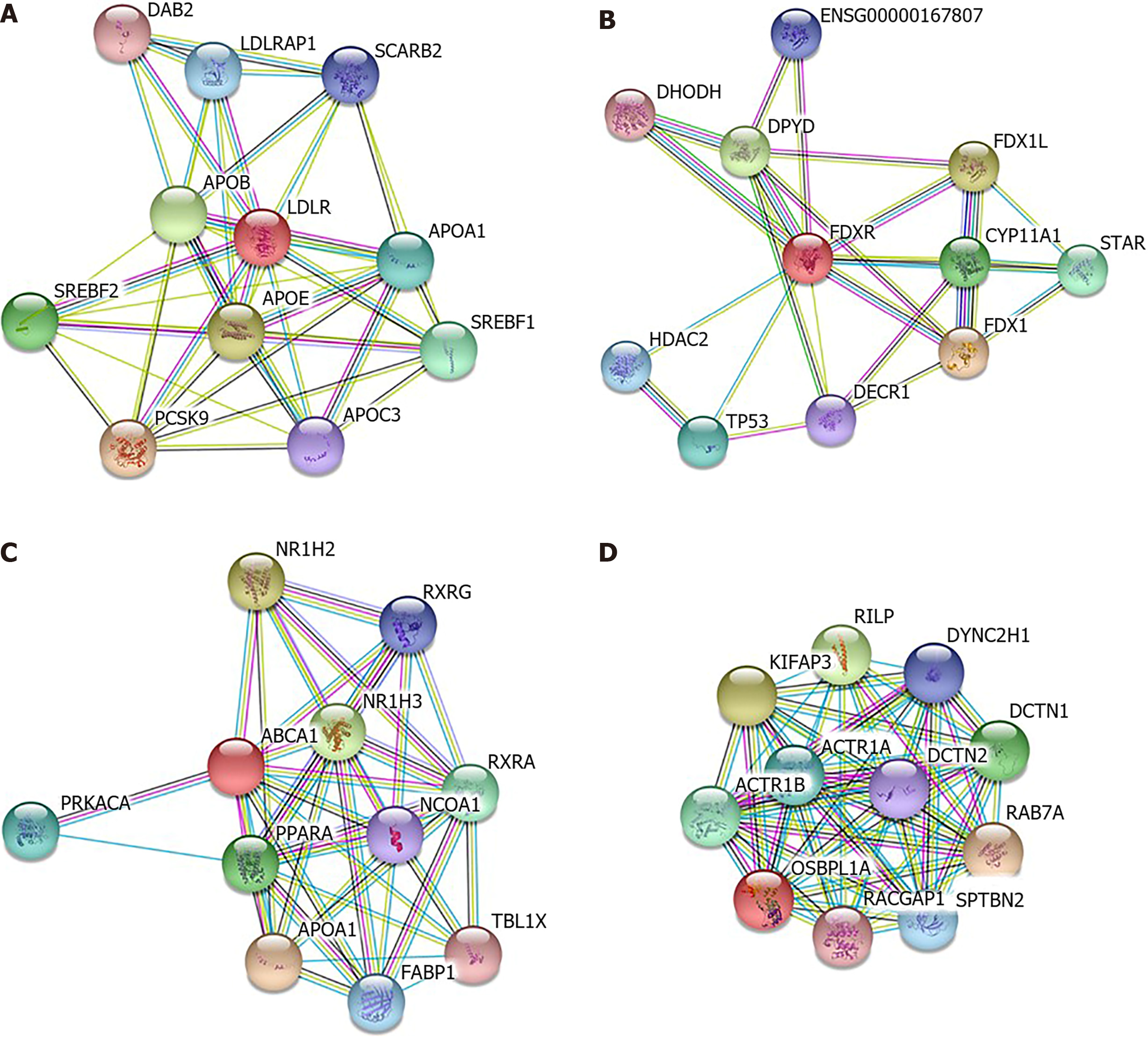
Figure 4 Protein–protein interaction network of differentially expressed genes with prognostic value.
Interacting nodes are displayed in colored circles using Search Tool for the Retrieval of Interacting Genes v10.0. A: Low-density lipoprotein receptor only involved in cholesterol metabolism pathway; B: FDXR was also involved in xenobiotic metabolic processes, cellular responses to xenobiotic stimuli and oxidation–reduction processes; C: ABCA1 was shown to be involved in the steroid hormone-mediated signaling pathway; D: OSBPL1A was involved in antigen processing and presentation of exogenous peptide antigens via MHC class II, microtubule-based movement and vesicle-mediated transport.
- Citation: Tao JH, Wang XT, Yuan W, Chen JN, Wang ZJ, Ma YB, Zhao FQ, Zhang LY, Ma J, Liu Q. Reduced serum high-density lipoprotein cholesterol levels and aberrantly expressed cholesterol metabolism genes in colorectal cancer. World J Clin Cases 2022; 10(14): 4446-4459
- URL: https://www.wjgnet.com/2307-8960/full/v10/i14/4446.htm
- DOI: https://dx.doi.org/10.12998/wjcc.v10.i14.4446