Published online Aug 12, 2015. doi: 10.5501/wjv.v4.i3.265
Peer-review started: January 27, 2015
First decision: March 6, 2015
Revised: March 23, 2015
Accepted: May 7, 2015
Article in press: May 8, 2015
Published online: August 12, 2015
Processing time: 200 Days and 10.6 Hours
Viruses are a cause of significant health problem worldwide, especially in the developing nations. Due to different anthropological activities, human populations are exposed to different viral pathogens, many of which emerge as outbreaks. In such situations, discovery of novel viruses is utmost important for deciding prevention and treatment strategies. Since last century, a number of different virus discovery methods, based on cell culture inoculation, sequence-independent PCR have been used for identification of a variety of viruses. However, the recent emergence and commercial availability of next-generation sequencers (NGS) has entirely changed the field of virus discovery. These massively parallel sequencing platforms can sequence a mixture of genetic materials from a very heterogeneous mix, with high sensitivity. Moreover, these platforms work in a sequence-independent manner, making them ideal tools for virus discovery. However, for their application in clinics, sample preparation or enrichment is necessary to detect low abundance virus populations. A number of techniques have also been developed for enrichment or viral nucleic acids. In this manuscript, we review the evolution of sequencing; NGS technologies available today as well as widely used virus enrichment technologies. We also discuss the challenges associated with their applications in the clinical virus discovery.
Core tip: Rapid development and commercial availability of next-generation sequencers (NGS) systems have dramatically changed almost every field of biological research, especially microbiology and metagenomics. Different NGS systems have been adapted and used for numerous applications in virology too. These systems are capable of rapidly sequencing and analyzing a complex mixture of nucleic acid templates, in a massively parallel fashion, making them ideal tools for viral metagenomics and discovery. This manuscript reviews the prevailing NGS technologies, their application in virus discovery to serve as a guide for the readers, working in the field of virology, public health and in biothreat mitigation programs.
- Citation: Datta S, Budhauliya R, Das B, Chatterjee S, Vanlalhmuaka, Veer V. Next-generation sequencing in clinical virology: Discovery of new viruses. World J Virology 2015; 4(3): 265-276
- URL: https://www.wjgnet.com/2220-3249/full/v4/i3/265.htm
- DOI: https://dx.doi.org/10.5501/wjv.v4.i3.265
Viral infections are a cause of significant health burden globally, particularly in the less developed countries. During the 20th century, methods for virus detection, characterization and taxonomical classification were established, that helped in the discovery of a number of important viruses, in prevention of viral infections and treatment. By the late-1950s, it was generally believed that most of the human pathogenic viruses had been discovered, but the emergence of a number of previously unknown viruses [Hepatitis viruses, Hantavirus, human immunodeficiency virus, Marburg virus, severe acute respiratory syndrome (SARS), Coronavirus Ebola virus] during the later part of the century strongly challenged this belief[1].
It has now become obvious that due to different anthropological activities, such as extensive globalization of travel and business, rapid unplanned urbanization, deforestation, etc., epidemiology of viral diseases have changed significantly[1]. This change has led to the increased exposure of different human populations to newer pathogens, including viruses, mostly zoonotic in nature[2,3]. The emergence of Ebola virus, Nipah virus, Sin Nombre Hantavirus, SARS, Influenza viruses (H1N1, H7N9), and MERS viruses in the recent past[4], clearly signify the onset of many others in the near future. According to a recent statistical estimate, there are at least 320,000 mammalian viruses that are waiting to be discovered[5]. The World Health Organization (WHO) has correctly cautioned that, “It would be extremely naïve and complacent to assume that there will be no other disease like AIDS, Ebola, or SARS, sooner or later”[6].
Apart from natural outbreaks, the risk of pathogens, especially deadly viruses, to be used as biological weapons and agents of bioterrorism have also increased in the recent years[7]. Being exceptionally diverse, in term of etiology, morphology, nucleic acid type and sequence information, clinical manifestations, etc., rapid detection and identification of viruses pose great challenge to clinical investigators. Nevertheless, during natural or deliberate outbreaks, identification and characterization of viruses in clinical samples is extremely essential to facilitate prevention and quarantine strategies, implement specific diagnostic tools, and also to determine explicit treatment strategy.
This article will review the gradual evolution and recent advances in the field of virus discovery, with special reference to the next-generation sequencing (NGS) technologies and related molecular biology methodologies.
Classically, virus discovery from clinical samples was based on filtration (to remove host cells and other microbes), inoculation of the cell free filtrate in suitable cell cultures followed by purification of the viruses from cultures and their characterization[8-10]. Morphological changes in the cultured cells, collectively known as cytopathic effect, such as formation of syncytia, cell rounding, lysis, detachment, or inclusion bodies, etc., indicate the presence and successful infection of the virus(es) in the cells[11]. Virus isolate(s) are purified from the cultured cells or culture supernatant using density gradient and other high speed centrifugal techniques. This is followed by structural characterization of viral particles, antigens, nucleic acids, through different biophysical and biochemical methods[4]. Although classical methods are sometimes considered as time-consuming, tedious and need significant experimental basis, but the cell inoculation method still remains an exceptional source of enriched viral particles required for serological, molecular characterization and other purposes. Nonetheless, in many cases, viruses are not readily infective to cell cultures, which severely hamper their characterization. Additionally, repeated passaging of the virus to obtain high titer could change the population of virus being sought[12].
Subsequently, with the development of nucleic acid sequence-dependent techniques, such as PCR-sequencing and microarrays, the requirement of cell culture based traditional methods became obsolete to a large extent[13-16]. These techniques were comparatively much faster as compared to classical techniques and led to the discovery of several new genotypes of known viruses. Among PCR and microarray based methods, the former gained enormous popularity due to its ability to rapidly amplify very small amounts of viral sequences from clinical samples. Even though, the prior requirement of sequence information (to design primers and hybridization probe), made this technique suitable for discovery of new genotypes of known viruses, but not appropriate for absolutely novel viruses. This limitation was later addressed by the development of consensus or degenerate PCR[17,18]. Although, this PCR method was tolerant to considerable sequence variation, but it lacked its original sensitivity and was still critically dependent on prior sequence information of the virus genera/family being investigated. Moreover, this method could only amplify small fractions of viral genome, which were sometimes not enough for further analysis.
The limitations of sequence dependent techniques prompted the investigators to resort to “metagenomics”, a technique that does not presume any knowledge about the organisms being investigated[19]. Metagenomics is the study of total genetic material present in a given sample, without culturing the organisms present in it. Conventional metagenomics analyses involved direct amplification of the nucleic acids through PCR, cloning and sequencing, etc.[15,20. At the outset, this technique was intensely used for assessing the bacterial diversity within highly diverse samples ranging from soil, oceans, and lakes to human gut and stool, which demonstrated the power of this technique to discover genetic materials of unknown origin[21]. Subsequently, early virus discovery investigators developed a number of random amplification techniques for viral metagenomics, such as sequence-independent single-primer amplification (SISPA), virus discovery based on cDNA-AFLP (VIDISCA), rolling circle amplification (RCA), etc., to amplify viral genetic materials for cloning and sequencing[15,20,22]. Extensive use of these viral metagenomic techniques, led to the discovery of different viruses, including human T-cell lymphotropic virus type-1, Torque Teno virus, different Parvoviruses, Coronaviruses, Polyomaviruses, Hepatitis C virus, Sin Nombre virus, Human Herpesviruses 6 and 8, and West Nile virus, etc., in clinical samples[23-26].
In all the above nucleic acid sequence based virus discovery approaches, the Sanger sequencing method played a very significant role. However, with the commercial availability of high throughput sequencing technologies in 2005, a gradual shift in generation of sequencing technologies became evident. These massively parallel sequencing technologies evolved rapidly and entirely transformed almost every field of biological research including clinical research laboratories[27]. NGS is presently the most attractive approach towards metagenomics, including viral metagenomics, due to its independence from the requirement of prior sequence information. Furthermore, being highly sensitive, NGS can rapidly recuperate nearly full genome sequences of viruses, with relatively less amount of starting material as compared to conventional cloning based approaches[28,29]. Moreover, the large dynamic detection range of the NGS has established it as the most powerful technology available till date, which has catalyzed the rate of virus discovery[30-33]. In combination with conventional methods such as SISPA, VIDISCA, RCA, etc., NGS can dramatically augment turnaround time and sensitivity of virus discovery[23]. Additionally, NGS has enormous, exciting applications in virology, including analysis of viral evolution and quasispecies analysis, antiviral resistance, vaccine, etc.[23,33-35].
A comparison of different virus discovery approaches, their advantages and limitations, applicability in different scenarios, etc., is presented in Table 1.
| Classical approaches(Cell culture and infection based) | Nucleic acid sequence-dependent amplification approaches | Nucleic acid sequence-independent amplification approaches | Next-generation sequencers-based metagenomic approaches | |
| Requirement of cell culture systems | Yes, required for virus particle enrichment | Not required | Not required | Not required |
| Information about the cytopathic effects of the virus | Yes, could be achieved through cell changes | No information could be achieved | No information could be achieved | No information could be achieved |
| Requirement of special equipments for purification | Yes, Ultracentrifuge/high speed centrifuges, density gradient is required for preparing pure virus | Not necessary, semi pure preparations obtained through low speed centrifuges are suitable | Not necessary, semi pure preparations obtained through low speed centrifuges are suitable | Not necessary, semi pure preparations obtained through low speed centrifuges are suitable |
| Information about detailed morphological/structural features of the virus | Yes, could be achieved through Electron/Atomic Force microscopy | No information on virus morphology/structure could be achieved directly | No information on virus morphology/structure could be achieved directly | No information on virus morphology/structure could be achieved directly |
| Time required for virus identification | Long time is required for identification, ranging from days to weeks | Comparatively faster, days required if cloning and sequencing is involved. Faster with microarray based approaches | Comparatively faster, virus could be identified within few days | Fastest available approach, identification could be done within days and even some times within hours |
| Requirement of prior knowledge about the virus | Not required | Some information is required regarding genus/family to design primers/probes | Being sequence independent technique, no information is required | Being sequence independent technique, no information is required |
| Dynamic detection range | Very narrow | Narrow | Wide | Extremely wide |
| Tolerance to non-viral materials | Vulnerable to other pathogens capable of infecting cell | Being sequence dependent, less vulnerable to other sequences from host and other pathogens | Being sequence in-dependent, more vulnerable to other sequences from host and other pathogens. Virus enrichment techniques required before analysis | Being sequence in-dependent, more vulnerable to other sequences from host and other pathogens. Virus enrichment techniques required before analysis |
| Suitability for discovery of new viruses | Yes | Less suitable, good at discovery of genotypes/variants of known viruses | Yes | Yes |
| Suitability during outbreaks | Not suitable due to requirement of long time | Not suitable due to requirement of prior sequence information | Yes, but still considerable time is required during outbreaks | Being fast, very much suitable in detecting pathogens in an outbreak scenario |
Originally, two different DNA sequencing methods were described almost simultaneously, the Sanger’s method, and the Maxam-Gilbert’s method[36,37], both considered as the first-generation of sequencing methods. Sanger’s method was based on DNA sequencing with chain-terminating inhibitors, while Maxam-Gilbert’s method was based on base-specific chemical modification and cleavage of the DNA backbone[38]. Due to its ease and possibility of automation, Sanger’s method became instantly popular and was successfully commercialized into DNA sequencing machines. As a result, for almost last 3 decades, the Sanger’s method dominated as the gold standard for DNA sequencing[39]. This sequencing method was primarily accomplished by amplification of templates with fluorescently labeled chain-terminating nucleotides, followed by capillary electrophoresis of the amplicons and reading the fluorescence signals, which can provide consistent sequence information of templates up to 1000 bp. Despite its wide use for sequencing pure templates, this sequence method was constrained by its low throughput, higher cost, time and labor involved in sequencing larger genomes. Furthermore, complete dependence on specific primers, inability to sequence the genetic material from a mix of diverse organisms severely restricted its use for direct metagenomic applications.
To overcome the technological constraints of the Sanger sequencers, second-generation or the NGS technologies were developed, based on a large number of innovations in the amplification technology, sequencing chemistry, microfluidics, imaging technologies, and Bioinformatics, etc.[40]. These novel sequencing technologies, initially commercialized by two companies, namely Roche and Illumina, and later by Life Technologies have spectacularly high throughput and high sensitivity, making them more appropriate for direct application in metagenomic studies. As compared to Sanger sequencers, currently available 2nd-generation NGS platforms are capable of generating only short sequence reads, but the true magnificence of NGS lies in their capability to sequence and analyze complex mixes of DNA in a massively parallel manner, generating millions to billions of sequence reads in a single run. Consequently, these technologies are often referred to as “short read” technologies and are distinguished by “third generation” sequencing technologies (or “long read”) that provide significantly longer reads (kilobases). However, at present, these long read technologies have, on the whole, lower throughput and accuracy[41-43].
Even though, widely distinct in their sequencing chemistry and detection technology, NGS platforms are common in terms of massively parallel sequencing of clonally amplified or single DNA molecules. On these platforms, sequencing is executed by repetitive cycles of polymerase-mediated nucleotide extension (Roche-454, Illumina GA) or oligonucleotide ligation (SOLiD). Using a “wash-and-scan” technique, sequence data is acquired as large sets of fluorescence or luminescence images of the flow-cell surface, subsequent to each repetitive sequencing cycle step[44]. This data is later compiled by using a computer-intensive pipeline for image integration, quality assessment, storage, processing and analysis. A typical NGS run generates several hundred megabases (Mb) to gigabases (Gb) of nucleotide sequence data, depending on the platform.
Although NGS platforms commercially available today, provide massive parallel sequencing, but due to their technological features and data output capabilities, every platform is suitable for certain specific applications. Hence, as per explicit requirements, NGS platform needs to be carefully selected. In cases of virus discovery, which is the scope of this review, NGS platforms capable of generating longer sequence reads are preferable over the others. Long reads are extremely useful for de novo read assembly and generation of longer contigs, which endow with improved statistical power of finding related sequences in nucleotide database searches[45]. Conversely, for characterization and analysis of virus variants and quasispecies, platforms providing high quality reads, i.e., less error and increased depth became the choice, over longer read lengths. In this review, we will discuss briefly the most popular NGS technologies (Illumina and Roche 454), widely used in virology. The details of the technologies, sequencing chemistries and other applications have been reviewed elsewhere in details[10,31,34].
The most widely used NGS is the Illumina sequencing technology, where clonal amplification of the template is attained to form DNA clusters, using primers attached to solid surface and sequencing is achieved via reversible dye-terminator technology. Although Illumina sequencing has higher sequence yield at a relatively low cost per base, this platform has a characteristic systematic base calling bias, exhibit differences in sequence quality, a higher sequencing error rate and increased single-base errors associated with GGC motifs[46-49].
On the other hand, 454 sequencing platforms are based on parallel pyrosequencing, utilizing sequencing-by-synthesis chemistry and chemiluminescence is detected to achieve nucleotide sequence. This method amplifies DNA through an emulsion PCR, generating clones of DNA using a single template. The main benefit of this technology is its ability to produce long reads, while restricted by its high error rate in homopolymers containing regions, and a high rate of artificial amplification[50-52]. The error rates of NGS are higher relative to the Sanger sequencers, and also require advanced computational tools and statistical calculations before further data processing and assembly[53]. Due to the NGS platform specific errors, presently, use of barcoding strategies, simultaneous sequencing of the samples by two different NGS platforms or high coverage sequencing have been recommended to counteract the effects of errors[54-56]. Nevertheless, these issues are being continually addressed and resolved in the newer versions of these platforms to make them more robust, both in terms of quality and quantity.
With the advancement in instrumentations, NGS platforms are now available as benchtop sequencing instruments in the form of the 454 GS Junior (Roche) and MiSeq (Illumina) which, despite having a small footprint, offer exciting NGS capabilities for clinical settings, at modest running costs[45]. MiSeq includes the Nextera, TruSeq, and reversible terminator-based sequencing by synthesis chemistry and has highest data integrity with broader range of application, including amplicon sequencing, clone checking, small genome sequencing etc. The MiSeq provides maximum throughput per run with lowest error rates, while the 454 GS Junior generates longer reads (approximately 600 bases) with better assemblies, but is limited by lower throughput and homopolymer-associated errors.
Apart from the two most widely used NGS technologies, another technology known as the SOLiD technology (by Life Technologies) is commercially available, but its representation in the scientific literature is limited compared to Roche 454 and Illumina, which might be attributable to its recent availability or complexity of data processing and assembly[57]. Nevertheless, SOLiD is slowly but gradually being accepted as a very reliable platform and has recently been used for de novo sequencing of a large mammalian genome[58].
Technical details of the NGS technologies have been extensively reviewed earlier[23,45,59]. A comparison of the currently available NSG systems is also available at the Genohub website (https://genohub.com/ngs-instrument-guide/).
The third generation of the sequencers has evolved lately, that include the Ion Torrent (Life Technologies), Single-Molecule Real-Time technology SMRT (Pacific Biosciences), and the Nanopore sequencing technology (Oxford Nanopore Technologies). Third-generation sequencers are distinct from their predecessors in two primary features: (1) template amplification is not needed prior to sequencing, which cuts down template preparation time; and cost (2) the signal is registered in real time, directly, during the enzymatic reaction. Apart from the Ion Torrent, rest of the third-generation sequencing technologies is quite recent, and still in the evaluation stages. Moreover, data on their application in the field of virus discovery is extremely scanty. Hence, all these will be discussed only briefly in this review.
The Ion Torrent Personal Genome Machine is based on a semiconductor based sequencing technology and does not require a fluorescence or chemiluminescence based image scanning, resulting in high speed, low cost sequencing system within small size equipment. Cyclically, the semiconductor microfluidic chip is flooded with each nucleotide, and a voltage is generated if it is incorporated, and no voltage is generated when not incorporated. This is based on the fact that every time a nucleotide is incorporated into the DNA molecules, a proton is released, causing a change in voltage, which is subsequently detected and registered by the chip[45,60].
Using the SMRT, single large DNA molecules can be sequenced with high processivity of up to 7 kb, with average read lengths of 3-4 kb[23,61]. On a SMRT cell, numerous Zero-Mode Waveguides are embedded with single set of enzymes and DNA template. During the reaction, enzyme incorporates a nucleotide into the complementary strand, cleaving off fluorescent dye linked with the nucleotide, and this fluorescent signal is captured[61].
Nanopore sequencing is another recently developed method of the third-generation sequencing[62,63]. Nanopore is a tiny biopore with diameter in nanoscale, and involves a heptameric transmembrane channel α-haemolysin (αHL) from Staphylococcus aureus. This protein has the ability to tolerate extraordinary voltage and current conditions (up to 100 mV, 100 pA). Under a standard condition of ionic flow, when a DNA molecule is passed through the channel of, etc., HL, current is modulated according to the size difference between every deoxyribonucleoside monophosphate (dNMP). This current modulation is detected by standard electrophysiological techniques and the dNMP is identified[62]. Nanopore sequencers are extremely small (size of a USB drive), can sequence long read faster (> 5 kb at a rate of 1 bp/ns), free of fluorescence/chemiluminescence and other enzymes, less sensitive to temperature and other conditions. These benefits make it fit as an extremely rapid sequencing device for field conditions, but the requirement of highly purified DNA needs to be addressed for their wide application in virus discovery.
Among the different NGS platforms available today, choosing the right one for correct application is extremely essential before embarking on a metagenomic project. In case of absence of a reference genome, or where highly divergent sequences are expected, such as in case of virus discovery, de novo sequencing and assembly is necessary. Such an assembly requires extensive computational power and datasets containing longer reads with higher coverage are preferable[64-66]. When reference genomes for assembly are available, technologies that generate short reads could also be used to have a high coverage of the metagenomes[53].
When compared in terms of publications, Illumina technology is the most widely used platform, irrespective of application. Earlier the use of this platform was not suited for virus discovery or de novo sequencing projects due to its short reads. However, regular augmentation in read length for Illumina platforms has made it suitable for de novo assembly of genomes, at a sensitivity, comparable to specific PCR[53,67,68]. However, according to the number of publications, specifically for metagenomic studies, pyrosequencing technology (Roche 454) is preferred over the other NGS approaches producing shorter reads. Of late, Roche has announced the discontinuation of its 454 technology by the mid-2016, which leaves the new investigators with alternative NGS platforms available today.
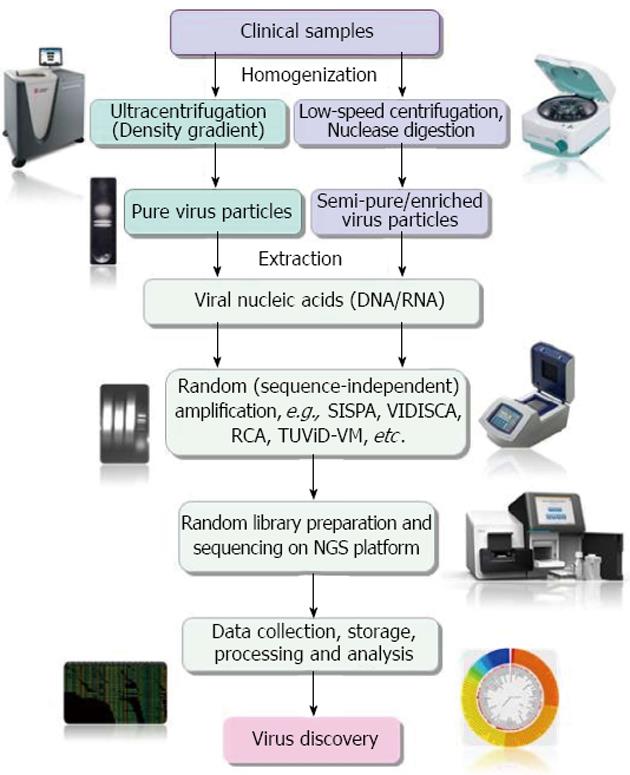
NGS has emerged as the most promising tool for the detection and discovery of novel infectious agents in clinical specimens[23]. However, being unbiased method of sequencing, NGS is greatly affected by very low virus-to-host genome ratios in clinical samples[69-71]. Hence, enrichment of pathogen genetic material or depletion of host genetic materials is essential to maximize sensitivity for discovery of novel pathogens, including viruses in clinical samples[23,72,73]. A schematic representation of the different steps involved in NGS based virus metagenomics and discovery is depicted in Figure 1.
A number of virus enrichment protocols involving physical and enzymatic techniques have been successfully applied for clinical samples. These include virus capsid purification through freeze/thaw cycles of cell disruption, filtration through appropriate pore membranes (0.45 μm and 0.22 μm), centrifugation, prior nuclease digestion of host genome, etc., followed by extraction of capsid-protected viral nucleic acids, their conversion to cDNA (in case of RNA virus) and non-specific PCR amplification[15]. The efficiency of enrichment in NGS-mediated virus discovery, especially the prior nuclease digestion has been clearly demonstrated by different studies[12,72,74]. Recently Hall et al[74] reviewed literatures available on methods for enrichment of viral nucleic acids from clinical samples for NGS-based studies. They found that both ultracentrifugation-mediated enrichment and low-speed centrifugation together with filtration and a nuclease digestion step is widely used for enrichment of viral nucleic acids.
Alternatively, approaches to deplete host genetic materials include use of methylation-specific DNase activity, host ribosomal RNA removal, duplex-specific nuclease normalization methods[75-77]. Such techniques on one hand increase the detection sensitivity of the NGS platform, circumventing the cost and time involved in generating and analyzing huge amounts of background data on the other hand. Ideally, in a clinical setting virus enrichment methods are required to be rapid, standardized and undemanding in terms of cost, manpower or instrumentation facility.
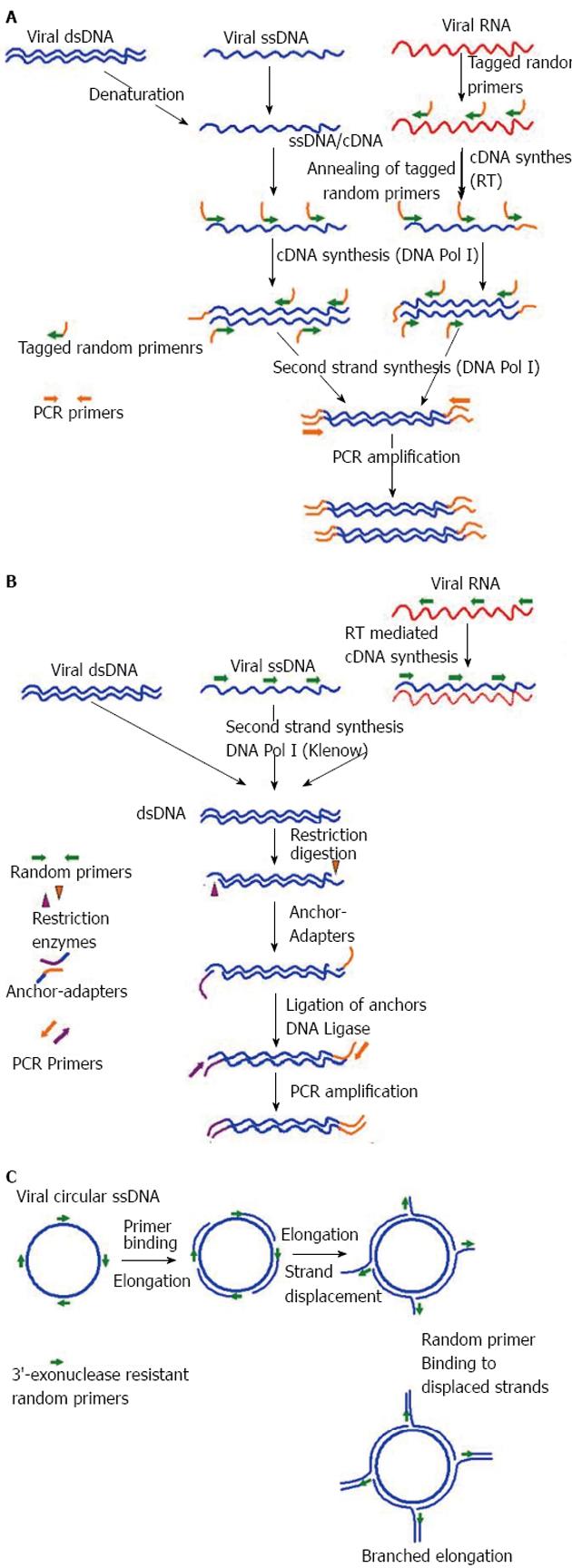
A number of virus enrichment methods have been applied successfully for NGS studies of different clinical samples. Of them, the sequence-independent single primer amplification (SISPA), developed by Reyes and Kim[78], was modified for successful amplification of viral sequences from serum by Allander et al[79] and later by others for identification of novel viruses through Sanger sequencing[80-83]. Recently, SISPA was used in combination with NGS and shown to be successful in detection of Hepatitis B and C viruses (HBV, HCV) in solid tissue samples[72]. In a recent study, SISPA-NGS strategy was found to be helpful in detection of Schmallenberg virus (SBV) in veterinary samples[84], suggesting the utility of this technique in screening of field animals that are intermediate hosts to many human viruses. In some of the recent studies no specific physical enrichment of virus particles was applied, but NGS was done on SISPA generated random PCR products, that also resulted in rapid detection of hemorrhagic fever-associated Yellow Fever Virus (YFV), Lujo virus (LUJV), and a new Arenavirus (related to lymphocytic choriomeningitis virus, LCMV) in diverse clinical samples[85-87].
Likewise, another well-established sequence-independent amplification technique is the virus discovery cDNA-amplified fragment length polymorphism (VIDISCA), used for discovery of a novel human SARS-associated coronavirus, HCoV-NL63[88,89]. Later this technique was successfully used in combination with Sanger sequencing to discover other novel viruses in clinical samples[90,91]. To late, the utility of this technique in combination with NGS for virus discovery has been demonstrated in veterinary samples, as well as in clinical samples[92,93]. Additionally, Shaukat et al[92] modified the VIDISCA method at the reverse transcription step by using specially designed mix of random hexamers that do not anneal to ribosomal RNA, further increasing the specificity of the assay. Apart from SISPA and VIDISCA, multiply-primed RCA has also been demonstrated to enrich circular viral genomes, suitable for sequencing through NSG platforms[94-96]. A diagrammatic representation of SISPA, VIDISCA and RCA is depicted in Figure 2 respectively. Recently, Kohl et al[97] reported an ultracentrifugation and DNA digestion based enrichment protocol followed by SISPA for detection of known and new viruses in human tissue samples. This technique, termed as tissue-based universal virus detection for viral metagenomics was demonstrated to complete within 28 h, making it suitable for discovery of zoonotic and biothreat agents of viral origin during outbreaks[97].
Alternatively, in another study, the authors used a barcoding strategy to carry out unbiased deep sequencing in multiple clinical samples and removed human and other low-quality sequences through bioinformatic filtering pipeline and identified viruses belonging to the Herpesviridae, Flaviviridae, Circoviridae, Anelloviridae, Asfarviridae, and Parvoviridae families in serum samples from tropical febrile illness[2].
Apart from virus discovery and detection in clinical samples, analysis of quasispecies, drug-resistant viral variants and monitoring of genetic consistency of live viral vaccines there are numerous applications of NGS, which are directly associated with human viral diseases. NGS-based virus detection technique has also been shown to be useful in surveillance of vector-borne and zoonotic viruses[23]. This possibility of detecting arthropod-borne viruses was demonstrated using Dengue virus-infected mosquito pools (Aedes aegypti), where, use of NGS resulted in highly sensitive detection of mosquito pools containing infected vectors[98]. Similarly, in a surveillance study focused on the discovery of bat-transmitted pathogens, using coronavirus consensus PCR and unbiased NGS, a new coronavirus related to SARS-CoV was documented[99].
Regardless of the field of applications and platforms used, ever-increasing capacities of NGS platforms and their wide usage have resulted in extremely unprecedented volumes of data. This is commonly referred to as “data deluge”, and is represented by huge NGS datasets deposited in specialized data archive such as the SRA, a primary archive of NIH, dedicated for submission and storage of raw data and alignment information, generated by all major NGS platforms. Being part of the International Nucleotide Sequence Database Collaboration at the National Center for Biotechnology Information, data submitted to either of the databases SRA, ENA (European nucleotide Archive of European Bioinformatics Institute, EBI) and the DDBJ (DNA Database of Japan) are shared amongst them. SRA serves as an initial point for downstream analysis of NGS data and also provide access to data from human clinical samples to authorized users. According to a recent comparison of GenBank statistics (Release 197, 8/2013 vs Release 203, 8/2014), total nucleotide entries to the GenBank represent an annual growth of more than 43%, and annual growth exclusively for virus sequence entries is 21%[100]. This data deluge has posed significant hardware, software and bioinformatics challenges towards storing, transfer, analysis and interpretation of the data[101].
All NGS platforms are advancing towards the capability to sequence longer DNA fragments, and to generate even larger volume of data sets[53]. To analyze such gigantic volumes of data, exceptionally massive computational facilities are also required, which has entirely revolutionized the field of Bioinformatics[60,102]. Once NGS sequence has been generated, the biggest of the challenges comes, i.e., computational requirements for storage and analysis of the massive data sets. Although a detailed description of bioinformatic processes involved in metagenomics data analysis is beyond the scale of this review, the key processes involved in the NGS data analysis are quality assessment, sequence assembly and annotation of the dataset against a database of nucleotide or protein sequences[34]. Quality assessment and data cleaning involves filtering out of low-quality sequences from the dataset, followed by alignment and error correction to separate true variance from the experimental noise[23]. After sequencing and quality assessment, there are two approaches for assembly of the reads. The sequence reads are then mapped to the available reference genome, or individual sequencing reads are assembled de novo, using different assembly servers[34,103]. The de novo approach is generally followed for discovery of viruses, considering the fact that reference genomes or related sequences may not be available in the databases. To determine the affinity of the assembled reads or the contigs, Basic Local Alignment Search Tool (BLAST) is used, that computes regions of similarity and statistical significance of possible matches between a query sequence and GenBank submissions[104]. Despite the availability of the BLAST, analyzing a viral metagenome may still be a challenging task in case of highly divergent or novel viral families, which are not represented in the database.
In the Table 2, we have summarized the challenges associated with handling and analysis of NGS generated data, their solutions presently available or suggested.
| Bioinformatics challenges associated with application of NGS in viral diagnostics | Action taken or proposed to overcome challenges |
| Generation of huge volumes of data by NGS platforms-“data deluge” | Advancement in storage and computation facilities, availability of computer with greater storage and highly powerful processors, cluster/grid computing and cloud computing. Computation facilities needs to be updated with emergence of newer platforms delivering larger datasets |
| Challenges in uploading data for submission to databases and supercomputing servers for analysis | Requirement of uninterrupted and extremely fast networks |
| Challenges in storage, public archival and ease of access | Creation of specialized data archive such as the Sequence Read Archive by NIH and ENA (European nucleotide Archive) by EBI. Sharing of data within the three major databases (NIH, EBI and DDBJ) for public accessibility |
| Challenges in analysis and visualization of large volumes of data, beyond the scope of computation facilities available in molecular biology laboratories | Creation of metagenomic or NGS data analysis pipelines and integrated tool kits, such as those available at NIH-NCBI, EMBL-EBI, MGRAST, CASAVA, MetaVir, Megan, UCSC Genome Browser, BioLinux, etc., availability of cloud computing based servers such as Galaxy |
| Challenges in alignment, de novo assembly, gene prediction and phylogenetic analyses NGS datasets, especially short read datasets | Availability of alignment algorithms/programs such as ABySS, ELAND, SOAP, Bowtie, Cloudburst, Zoom, BWA, SHRiMP, MOM, SeqMap, Metagene, Velvet, QSRA, ALLPATHS, EDENA, VCAKE, FragGeneScan, BLAST, GLIMMER, EULER-SR, Avadis, Eagle View, etc. |
| Interpretation of huge amount of data generated in metagenomic analyses by NGS platforms | Proper interpretation of analyzed data is of utmost importance to identify newer pathogens as well as their clinical significance |
During the last decade, numerous innovations in virus enrichment techniques, sequencing chemistry and signal detection technologies, availability of high end dedicated bioinformatic servers for analysis of the NGS data has greatly accelerated the discovery of viral pathogens in clinical samples. Apart from its increasing applications in virus discovery, NGS has been successfully used in monitoring of antiviral drug resistance, investigation of viral evolution, diversity and quasispecies, and evaluation of the human virome. The supreme advantage of the NGS platforms is their ability to characterize hundreds of different pathogens simultaneously that are not otherwise cultivable using conventional approaches. Nevertheless, there are a number of challenges that need to be overcome for these technologies to become routine in clinical settings. The initial cost of set-up, turnaround time, requirement of powerful computational facilities along with the requirement of a highly skilled group of people are the major barriers to their wide application in resource-limited countries, where the cases of emerging viruses are the highest.
Despite the broad utility of NGS in virus discovery, extremely high sensitivity of this technique also makes it prone to unintentional contamination. The use of random primers for enrichment and the deep sequencing may result in significant potential for carryover contamination from laboratory reagents. Simultaneous analyses of blinded controls may be one approach towards excluding such possibilities, but it will also double the cost of sequencing. Another outcome of the NGS data is the rapid rate of discovery of viruses. However, the absence of appropriate cell culture systems or animal models limit the possibility of experimental studies on these new viruses, thereby the clinical significance of these new viruses remains to be properly understood.
We thankfully acknowledge the Defence Research and Development Organization (DRDO), Ministry of Defence, Government of India for funding and support. We also thank the editor and three anonymous reviewers for their constructive comments, which helped us immensely to improve this manuscript.
P- Reviewer: Chen YD, Demonacos C, Qiu HJ S- Editor: Song XX L- Editor: A E- Editor: Yan JL
| 1. | Bichaud L, de Lamballerie X, Alkan C, Izri A, Gould EA, Charrel RN. Arthropods as a source of new RNA viruses. Microb Pathog. 2014;77:136-141. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 17] [Article Influence: 1.7] [Reference Citation Analysis (1)] |
| 2. | Yozwiak NL, Skewes-Cox P, Stenglein MD, Balmaseda A, Harris E, DeRisi JL. Virus identification in unknown tropical febrile illness cases using deep sequencing. PLoS Negl Trop Dis. 2012;6:e1485. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 134] [Cited by in F6Publishing: 138] [Article Influence: 11.5] [Reference Citation Analysis (1)] |
| 3. | Jones KE, Patel NG, Levy MA, Storeygard A, Balk D, Gittleman JL, Daszak P. Global trends in emerging infectious diseases. Nature. 2008;451:990-993. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4341] [Cited by in F6Publishing: 4297] [Article Influence: 268.6] [Reference Citation Analysis (0)] |
| 4. | Dong J, Olano JP, McBride JW, Walker DH. Emerging pathogens: challenges and successes of molecular diagnostics. J Mol Diagn. 2008;10:185-197. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 88] [Cited by in F6Publishing: 92] [Article Influence: 5.8] [Reference Citation Analysis (2)] |
| 5. | Anthony SJ, Epstein JH, Murray KA, Navarrete-Macias I, Zambrana-Torrelio CM, Solovyov A, Ojeda-Flores R, Arrigo NC, Islam A, Ali Khan S. A strategy to estimate unknown viral diversity in mammals. MBio. 2013;4:e00598-e00513. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 279] [Cited by in F6Publishing: 245] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 6. | World Health Annual Report 2007. [accessed 2015 Jan 1]. Available from: http://www.who.int/whr/2007/overview/en/index2.html. [Cited in This Article: ] |
| 7. | Bronze MS, Huycke MM, Machado LJ, Voskuhl GW, Greenfield RA. Viral agents as biological weapons and agents of bioterrorism. Am J Med Sci. 2002;323:316-325. [PubMed] [Cited in This Article: ] |
| 8. | Todaro GJ, Zeve V, Aaronson SA. Cell culture techniques in the search for cancer viruses of man. In Vitro. 1971;6:355-361. [PubMed] [Cited in This Article: ] |
| 9. | Herrmann EC. New concepts and developments in applied diagnostic virology. Prog Med Virol. 1974;17:221-289. [PubMed] [Cited in This Article: ] |
| 10. | Lipkin WI, Firth C. Viral surveillance and discovery. Curr Opin Virol. 2013;3:199-204. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in F6Publishing: 42] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 11. | Friedman RM, Ramseur JM. Mechanisms of persistent infections by cytopathic viruses in tissue culture. Brief review. Arch Virol. 1979;60:83-103. [PubMed] [Cited in This Article: ] |
| 12. | Neill JD, Bayles DO, Ridpath JF. Simultaneous rapid sequencing of multiple RNA virus genomes. J Virol Methods. 2014;201:68-72. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 57] [Article Influence: 5.7] [Reference Citation Analysis (2)] |
| 13. | Haagmans BL, Andeweg AC, Osterhaus AD. The application of genomics to emerging zoonotic viral diseases. PLoS Pathog. 2009;5:e1000557. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 44] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 14. | Ansorge WJ. Next-generation DNA sequencing techniques. N Biotechnol. 2009;25:195-203. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 600] [Cited by in F6Publishing: 470] [Article Influence: 31.3] [Reference Citation Analysis (0)] |
| 15. | Delwart EL. Viral metagenomics. Rev Med Virol. 2007;17:115-131. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 230] [Cited by in F6Publishing: 224] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 16. | Wang D, Coscoy L, Zylberberg M, Avila PC, Boushey HA, Ganem D, DeRisi JL. Microarray-based detection and genotyping of viral pathogens. Proc Natl Acad Sci USA. 2002;99:15687-15692. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 594] [Cited by in F6Publishing: 540] [Article Influence: 24.5] [Reference Citation Analysis (0)] |
| 17. | Rose TM. CODEHOP-mediated PCR - a powerful technique for the identification and characterization of viral genomes. Virol J. 2005;2:20. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 56] [Cited by in F6Publishing: 63] [Article Influence: 3.3] [Reference Citation Analysis (1)] |
| 18. | Kricka LJ. Nucleic acid detection technologies -- labels, strategies, and formats. Clin Chem. 1999;45:453-458. [PubMed] [Cited in This Article: ] |
| 19. | Handelsman J. Metagenomics: application of genomics to uncultured microorganisms. Microbiol Mol Biol Rev. 2004;68:669-685. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1529] [Cited by in F6Publishing: 1147] [Article Influence: 60.4] [Reference Citation Analysis (0)] |
| 20. | Bexfield N, Kellam P. Metagenomics and the molecular identification of novel viruses. Vet J. 2011;190:191-198. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 65] [Cited by in F6Publishing: 63] [Article Influence: 4.5] [Reference Citation Analysis (1)] |
| 21. | Schloss PD, Handelsman J. Metagenomics for studying unculturable microorganisms: cutting the Gordian knot. Genome Biol. 2005;6:229. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 295] [Cited by in F6Publishing: 218] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 22. | Thurber RV, Haynes M, Breitbart M, Wegley L, Rohwer F. Laboratory procedures to generate viral metagenomes. Nat Protoc. 2009;4:470-483. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 417] [Cited by in F6Publishing: 407] [Article Influence: 27.1] [Reference Citation Analysis (0)] |
| 23. | Barzon L, Lavezzo E, Militello V, Toppo S, Palù G. Applications of next-generation sequencing technologies to diagnostic virology. Int J Mol Sci. 2011;12:7861-7884. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 191] [Cited by in F6Publishing: 177] [Article Influence: 13.6] [Reference Citation Analysis (2)] |
| 24. | Chiu CY. Viral pathogen discovery. Curr Opin Microbiol. 2013;16:468-478. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 151] [Cited by in F6Publishing: 146] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 25. | Schelhorn SE. Going viral- An integrated view on virological data analysis from basic research to clinical applications. PhD Dissertation. 2013, Saarland University, Germany. Available from: http://d-nb.info/1053980728/34. [Cited in This Article: ] |
| 26. | Boheemen SV. Virus Discovery and Characterization using Next-Generation Sequencing. PhD Dissertation. 2014, Erasmus Medical Center, Rotterdam. Available from: http://repub.eur.nl/pub/76063/. [Cited in This Article: ] |
| 27. | Lecuit M, Eloit M. The human virome: new tools and concepts. Trends Microbiol. 2013;21:510-515. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 89] [Cited by in F6Publishing: 85] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 28. | Marston DA, McElhinney LM, Ellis RJ, Horton DL, Wise EL, Leech SL, David D, de Lamballerie X, Fooks AR. Next generation sequencing of viral RNA genomes. BMC Genomics. 2013;14:444. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 110] [Cited by in F6Publishing: 113] [Article Influence: 10.3] [Reference Citation Analysis (1)] |
| 29. | Boonham N, Kreuze J, Winter S, van der Vlugt R, Bergervoet J, Tomlinson J, Mumford R. Methods in virus diagnostics: from ELISA to next generation sequencing. Virus Res. 2014;186:20-31. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 240] [Cited by in F6Publishing: 169] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 30. | Wang D. Fruits of virus discovery: new pathogens and new experimental models. J Virol. 2015;89:1486-1488. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 8] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 31. | Capobianchi MR, Giombini E, Rozera G. Next-generation sequencing technology in clinical virology. Clin Microbiol Infect. 2013;19:15-22. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 125] [Cited by in F6Publishing: 117] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 32. | Moore RA, Warren RL, Freeman JD, Gustavsen JA, Chénard C, Friedman JM, Suttle CA, Zhao Y, Holt RA. The sensitivity of massively parallel sequencing for detecting candidate infectious agents associated with human tissue. PLoS One. 2011;6:e19838. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 51] [Cited by in F6Publishing: 53] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 33. | Svraka S, Rosario K, Duizer E, van der Avoort H, Breitbart M, Koopmans M. Metagenomic sequencing for virus identification in a public-health setting. J Gen Virol. 2010;91:2846-2856. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 48] [Article Influence: 3.4] [Reference Citation Analysis (1)] |
| 34. | Radford AD, Chapman D, Dixon L, Chantrey J, Darby AC, Hall N. Application of next-generation sequencing technologies in virology. J Gen Virol. 2012;93:1853-1868. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 187] [Cited by in F6Publishing: 179] [Article Influence: 14.9] [Reference Citation Analysis (1)] |
| 35. | Lipkin WI. Microbe hunting. Microbiol Mol Biol Rev. 2010;74:363-377. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 91] [Cited by in F6Publishing: 95] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 36. | Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463-5467. [PubMed] [Cited in This Article: ] |
| 37. | Maxam AM, Gilbert W. A new method for sequencing DNA. 1977. Biotechnology. 1992;24:99-103. [PubMed] [Cited in This Article: ] |
| 38. | Friedmann T. Rapid nucleotide sequencing of DNA. Am J Hum Genet. 1979;31:19-28. [PubMed] [Cited in This Article: ] |
| 39. | Grada A, Weinbrecht K. Next-generation sequencing: methodology and application. J Invest Dermatol. 2013;133:e11. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 171] [Cited by in F6Publishing: 138] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 40. | Buermans HP, den Dunnen JT. Next generation sequencing technology: Advances and applications. Biochim Biophys Acta. 2014;1842:1932-1941. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 435] [Cited by in F6Publishing: 460] [Article Influence: 46.0] [Reference Citation Analysis (0)] |
| 41. | Flaherty P, Natsoulis G, Muralidharan O, Winters M, Buenrostro J, Bell J, Brown S, Holodniy M, Zhang N, Ji HP. Ultrasensitive detection of rare mutations using next-generation targeted resequencing. Nucleic Acids Res. 2012;40:e2. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 102] [Cited by in F6Publishing: 103] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 42. | Xu X, Hou Y, Yin X, Bao L, Tang A, Song L, Li F, Tsang S, Wu K, Wu H. Single-cell exome sequencing reveals single-nucleotide mutation characteristics of a kidney tumor. Cell. 2012;148:886-895. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 513] [Cited by in F6Publishing: 487] [Article Influence: 40.6] [Reference Citation Analysis (0)] |
| 43. | Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, Cook K, Stepansky A, Levy D, Esposito D. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90-94. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1880] [Cited by in F6Publishing: 1866] [Article Influence: 143.5] [Reference Citation Analysis (0)] |
| 44. | Schadt EE, Turner S, Kasarskis A. A window into third-generation sequencing. Hum Mol Genet. 2010;19:R227-R240. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 611] [Cited by in F6Publishing: 636] [Article Influence: 45.4] [Reference Citation Analysis (0)] |
| 45. | Loman NJ, Misra RV, Dallman TJ, Constantinidou C, Gharbia SE, Wain J, Pallen MJ. Performance comparison of benchtop high-throughput sequencing platforms. Nat Biotechnol. 2012;30:434-439. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 999] [Cited by in F6Publishing: 1045] [Article Influence: 87.1] [Reference Citation Analysis (0)] |
| 46. | Nakamura K, Oshima T, Morimoto T, Ikeda S, Yoshikawa H, Shiwa Y, Ishikawa S, Linak MC, Hirai A, Takahashi H. Sequence-specific error profile of Illumina sequencers. Nucleic Acids Res. 2011;39:e90. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 434] [Cited by in F6Publishing: 411] [Article Influence: 31.6] [Reference Citation Analysis (0)] |
| 47. | Schröder J, Bailey J, Conway T, Zobel J. Reference-free validation of short read data. PLoS One. 2010;5:e12681. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 22] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 48. | Erlich Y, Mitra PP, delaBastide M, McCombie WR, Hannon GJ. Alta-Cyclic: a self-optimizing base caller for next-generation sequencing. Nat Methods. 2008;5:679-682. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 135] [Cited by in F6Publishing: 134] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 49. | Dolan PC, Denver DR. TileQC: a system for tile-based quality control of Solexa data. BMC Bioinformatics. 2008;9:250. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 29] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 50. | Quince C, Lanzén A, Curtis TP, Davenport RJ, Hall N, Head IM, Read LF, Sloan WT. Accurate determination of microbial diversity from 454 pyrosequencing data. Nat Methods. 2009;6:639-641. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 792] [Cited by in F6Publishing: 703] [Article Influence: 46.9] [Reference Citation Analysis (0)] |
| 51. | Gomez-Alvarez V, Teal TK, Schmidt TM. Systematic artifacts in metagenomes from complex microbial communities. ISME J. 2009;3:1314-1317. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 354] [Cited by in F6Publishing: 329] [Article Influence: 21.9] [Reference Citation Analysis (0)] |
| 52. | Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376-380. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5541] [Cited by in F6Publishing: 4999] [Article Influence: 263.1] [Reference Citation Analysis (0)] |
| 53. | Escalante AE, Lev JardŦn Barbolla, Santiago RamØrez-Barahona, Luis E. Eguiarte. The study of biodiversity in the era of massive sequencing. Rev Mex de Biodivers. 2014;85:1249-1264. [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 8] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 54. | Fox EJ, Bayliss KSR, Emond MJ, Loeb LA. Accuracy of Next Generation Sequencing Platforms. Next Generat Sequenc Applic. 2014;1:1. [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 70] [Reference Citation Analysis (0)] |
| 55. | Dalloul RA, Long JA, Zimin AV, Aslam L, Beal K, Blomberg Le Ann, Bouffard P, Burt DW, Crasta O, Crooijmans RP. Multi-platform next-generation sequencing of the domestic turkey (Meleagris gallopavo): genome assembly and analysis. PLoS Biol. 2010;8:pii: e1000475. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 316] [Cited by in F6Publishing: 320] [Article Influence: 22.9] [Reference Citation Analysis (0)] |
| 56. | Aury JM, Cruaud C, Barbe V, Rogier O, Mangenot S, Samson G, Poulain J, Anthouard V, Scarpelli C, Artiguenave F. High quality draft sequences for prokaryotic genomes using a mix of new sequencing technologies. BMC Genomics. 2008;9:603. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 66] [Cited by in F6Publishing: 73] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 57. | Flicek P, Birney E. Sense from sequence reads: methods for alignment and assembly. Nat Methods. 2009;6:S6-S12. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 251] [Cited by in F6Publishing: 266] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 58. | Rubin CJ, Megens HJ, Martinez Barrio A, Maqbool K, Sayyab S, Schwochow D, Wang C, Carlborg Ö, Jern P, Jørgensen CB. Strong signatures of selection in the domestic pig genome. Proc Natl Acad Sci USA. 2012;109:19529-19536. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 389] [Cited by in F6Publishing: 431] [Article Influence: 35.9] [Reference Citation Analysis (0)] |
| 59. | Metzker ML. Sequencing technologies - the next generation. Nat Rev Genet. 2010;11:31-46. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4795] [Cited by in F6Publishing: 4025] [Article Influence: 268.3] [Reference Citation Analysis (0)] |
| 60. | Mardis ER. Next-generation DNA sequencing methods. Annu Rev Genomics Hum Genet. 2008;9:387-402. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1467] [Cited by in F6Publishing: 1182] [Article Influence: 73.9] [Reference Citation Analysis (0)] |
| 61. | Quail MA, Smith M, Coupland P, Otto TD, Harris SR, Connor TR, Bertoni A, Swerdlow HP, Gu Y. A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genomics. 2012;13:341. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1386] [Cited by in F6Publishing: 1225] [Article Influence: 102.1] [Reference Citation Analysis (1)] |
| 62. | Branton D, Deamer DW, Marziali A, Bayley H, Benner SA, Butler T, Di Ventra M, Garaj S, Hibbs A, Huang X. The potential and challenges of nanopore sequencing. Nat Biotechnol. 2008;26:1146-1153. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1870] [Cited by in F6Publishing: 1603] [Article Influence: 100.2] [Reference Citation Analysis (0)] |
| 63. | Laszlo AH, Derrington IM, Ross BC, Brinkerhoff H, Adey A, Nova IC, Craig JM, Langford KW, Samson JM, Daza R. Decoding long nanopore sequencing reads of natural DNA. Nat Biotechnol. 2014;32:829-833. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 341] [Cited by in F6Publishing: 292] [Article Influence: 29.2] [Reference Citation Analysis (0)] |
| 64. | Cahais V, Gayral P, Tsagkogeorga G, Melo-Ferreira J, Ballenghien M, Weinert L, Chiari Y, Belkhir K, Ranwez V, Galtier N. Reference-free transcriptome assembly in non-model animals from next-generation sequencing data. Mol Ecol Resour. 2012;12:834-845. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 2] [Reference Citation Analysis (0)] |
| 65. | Glenn TC. Field guide to next-generation DNA sequencers. Mol Ecol Resour. 2011;11:759-769. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 782] [Cited by in F6Publishing: 642] [Article Influence: 49.4] [Reference Citation Analysis (0)] |
| 66. | Martin JA, Wang Z. Next-generation transcriptome assembly. Nat Rev Genet. 2011;12:671-682. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 860] [Cited by in F6Publishing: 895] [Article Influence: 68.8] [Reference Citation Analysis (0)] |
| 67. | Shokralla S, Spall JL, Gibson JF, Hajibabaei M. Next-generation sequencing technologies for environmental DNA research. Mol Ecol. 2012;21:1794-1805. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 582] [Cited by in F6Publishing: 593] [Article Influence: 49.4] [Reference Citation Analysis (0)] |
| 68. | Cheval J, Sauvage V, Frangeul L, Dacheux L, Guigon G, Dumey N, Pariente K, Rousseaux C, Dorange F, Berthet N. Evaluation of high-throughput sequencing for identifying known and unknown viruses in biological samples. J Clin Microbiol. 2011;49:3268-3275. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 117] [Cited by in F6Publishing: 119] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 69. | He B, Li Z, Yang F, Zheng J, Feng Y, Guo H, Li Y, Wang Y, Su N, Zhang F. Virome profiling of bats from Myanmar by metagenomic analysis of tissue samples reveals more novel Mammalian viruses. PLoS One. 2013;8:e61950. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 90] [Cited by in F6Publishing: 96] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 70. | Baker KS, Leggett RM, Bexfield NH, Alston M, Daly G, Todd S, Tachedjian M, Holmes CE, Crameri S, Wang LF. Metagenomic study of the viruses of African straw-coloured fruit bats: detection of a chiropteran poxvirus and isolation of a novel adenovirus. Virology. 2013;441:95-106. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 111] [Cited by in F6Publishing: 107] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 71. | Nakamura S, Yang CS, Sakon N, Ueda M, Tougan T, Yamashita A, Goto N, Takahashi K, Yasunaga T, Ikuta K. Direct metagenomic detection of viral pathogens in nasal and fecal specimens using an unbiased high-throughput sequencing approach. PLoS One. 2009;4:e4219. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 194] [Cited by in F6Publishing: 174] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 72. | Daly GM, Bexfield N, Heaney J, Stubbs S, Mayer AP, Palser A, Kellam P, Drou N, Caccamo M, Tiley L. A viral discovery methodology for clinical biopsy samples utilising massively parallel next generation sequencing. PLoS One. 2011;6:e28879. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 50] [Cited by in F6Publishing: 58] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 73. | Whon TW, Kim MS, Roh SW, Shin NR, Lee HW, Bae JW. Metagenomic characterization of airborne viral DNA diversity in the near-surface atmosphere. J Virol. 2012;86:8221-8231. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 87] [Cited by in F6Publishing: 82] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 74. | Hall RJ, Wang J, Todd AK, Bissielo AB, Yen S, Strydom H, Moore NE, Ren X, Huang QS, Carter PE. Evaluation of rapid and simple techniques for the enrichment of viruses prior to metagenomic virus discovery. J Virol Methods. 2014;195:194-204. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 137] [Cited by in F6Publishing: 144] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 75. | Oyola SO, Gu Y, Manske M, Otto TD, O’Brien J, Alcock D, Macinnis B, Berriman M, Newbold CI, Kwiatkowski DP. Efficient depletion of host DNA contamination in malaria clinical sequencing. J Clin Microbiol. 2013;51:745-751. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 60] [Cited by in F6Publishing: 62] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 76. | He S, Wurtzel O, Singh K, Froula JL, Yilmaz S, Tringe SG, Wang Z, Chen F, Lindquist EA, Sorek R. Validation of two ribosomal RNA removal methods for microbial metatranscriptomics. Nat Methods. 2010;7:807-812. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 164] [Cited by in F6Publishing: 168] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 77. | Shagina I, Bogdanova E, Mamedov IZ, Lebedev Y, Lukyanov S, Shagin D. Normalization of genomic DNA using duplex-specific nuclease. Biotechniques. 2010;48:455-459. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 31] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 78. | Reyes GR, Kim JP. Sequence-independent, single-primer amplification (SISPA) of complex DNA populations. Mol Cell Probes. 1991;5:473-481. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 114] [Cited by in F6Publishing: 123] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 79. | Allander T, Emerson SU, Engle RE, Purcell RH, Bukh J. A virus discovery method incorporating DNase treatment and its application to the identification of two bovine parvovirus species. Proc Natl Acad Sci USA. 2001;98:11609-11614. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 297] [Cited by in F6Publishing: 296] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 80. | Cheng WX, Li JS, Huang CP, Yao DP, Liu N, Cui SX, Jin Y, Duan ZJ. Identification and nearly full-length genome characterization of novel porcine bocaviruses. PLoS One. 2010;5:e13583. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 91] [Cited by in F6Publishing: 94] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 81. | Abed Y, Boivin G. Molecular characterization of viruses from clinical respiratory samples producing unidentified cytopathic effects in cell culture. Viruses. 2009;1:84-90. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 5] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 82. | Victoria JG, Kapoor A, Li L, Blinkova O, Slikas B, Wang C, Naeem A, Zaidi S, Delwart E. Metagenomic analyses of viruses in stool samples from children with acute flaccid paralysis. J Virol. 2009;83:4642-4651. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 308] [Cited by in F6Publishing: 305] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 83. | Kirkland PD, Frost MJ, Finlaison DS, King KR, Ridpath JF, Gu X. Identification of a novel virus in pigs--Bungowannah virus: a possible new species of pestivirus. Virus Res. 2007;129:26-34. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 119] [Cited by in F6Publishing: 121] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 84. | Rosseel T, Scheuch M, Höper D, De Regge N, Caij AB, Vandenbussche F, Van Borm S. DNase SISPA-next generation sequencing confirms Schmallenberg virus in Belgian field samples and identifies genetic variation in Europe. PLoS One. 2012;7:e41967. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 44] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 85. | McMullan LK, Frace M, Sammons SA, Shoemaker T, Balinandi S, Wamala JF, Lutwama JJ, Downing RG, Stroeher U, MacNeil A. Using next generation sequencing to identify yellow fever virus in Uganda. Virology. 2012;422:1-5. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 41] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 86. | Briese T, Paweska JT, McMullan LK, Hutchison SK, Street C, Palacios G, Khristova ML, Weyer J, Swanepoel R, Egholm M. Genetic detection and characterization of Lujo virus, a new hemorrhagic fever-associated arenavirus from southern Africa. PLoS Pathog. 2009;5:e1000455. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 363] [Cited by in F6Publishing: 325] [Article Influence: 21.7] [Reference Citation Analysis (0)] |
| 87. | Palacios G, Druce J, Du L, Tran T, Birch C, Briese T, Conlan S, Quan PL, Hui J, Marshall J. A new arenavirus in a cluster of fatal transplant-associated diseases. N Engl J Med. 2008;358:991-998. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 541] [Cited by in F6Publishing: 476] [Article Influence: 29.8] [Reference Citation Analysis (0)] |
| 88. | Pyrc K, Jebbink MF, Berkhout B, van der Hoek L. Detection of new viruses by VIDISCA. Virus discovery based on cDNA-amplified fragment length polymorphism. Methods Mol Biol. 2008;454:73-89. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 23] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 89. | van der Hoek L, Pyrc K, Jebbink MF, Vermeulen-Oost W, Berkhout RJ, Wolthers KC, Wertheim-van Dillen PM, Kaandorp J, Spaargaren J, Berkhout B. Identification of a new human coronavirus. Nat Med. 2004;10:368-373. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1250] [Cited by in F6Publishing: 1265] [Article Influence: 63.3] [Reference Citation Analysis (0)] |
| 90. | de Souza Luna LK, Baumgarte S, Grywna K, Panning M, Drexler JF, Drosten C. Identification of a contemporary human parechovirus type 1 by VIDISCA and characterisation of its full genome. Virol J. 2008;5:26. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 26] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 91. | de Vries M, Pyrc K, Berkhout R, Vermeulen-Oost W, Dijkman R, Jebbink MF, Bruisten S, Berkhout B, van der Hoek L. Human parechovirus type 1, 3, 4, 5, and 6 detection in picornavirus cultures. J Clin Microbiol. 2008;46:759-762. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in F6Publishing: 47] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 92. | Shaukat S, Angez M, Alam MM, Jebbink MF, Deijs M, Canuti M, Sharif S, de Vries M, Khurshid A, Mahmood T. Identification and characterization of unrecognized viruses in stool samples of non-polio acute flaccid paralysis children by simplified VIDISCA. Virol J. 2014;11:146. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 17] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 93. | van der Heijden M, de Vries M, van Steenbeek FG, Favier RP, Deijs M, Brinkhof B, Rothuizen J, van der Hoek L, Penning LC. Sequence-independent VIDISCA-454 technique to discover new viruses in canine livers. J Virol Methods. 2012;185:152-155. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 10] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 94. | de Vries M, Deijs M, Canuti M, van Schaik BD, Faria NR, van de Garde MD, Jachimowski LC, Jebbink MF, Jakobs M, Luyf AC. A sensitive assay for virus discovery in respiratory clinical samples. PLoS One. 2011;6:e16118. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 73] [Cited by in F6Publishing: 74] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 95. | Meiring TL, Salimo AT, Coetzee B, Maree HJ, Moodley J, Hitzeroth II, Freeborough MJ, Rybicki EP, Williamson AL. Next-generation sequencing of cervical DNA detects human papillomavirus types not detected by commercial kits. Virol J. 2012;9:164. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 54] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 96. | Sijmons S, Thys K, Corthout M, Van Damme E, Van Loock M, Bollen S, Baguet S, Aerssens J, Van Ranst M, Maes P. A method enabling high-throughput sequencing of human cytomegalovirus complete genomes from clinical isolates. PLoS One. 2014;9:e95501. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 22] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 97. | Kohl C, Brinkmann A, Dabrowski PW, Radonić A, Nitsche A, Kurth A. Protocol for metagenomic virus detection in clinical specimens. Emerg Infect Dis. 2015;21:48-57. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 72] [Cited by in F6Publishing: 76] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 98. | Bishop-Lilly KA, Turell MJ, Willner KM, Butani A, Nolan NM, Lentz SM, Akmal A, Mateczun A, Brahmbhatt TN, Sozhamannan S. Arbovirus detection in insect vectors by rapid, high-throughput pyrosequencing. PLoS Negl Trop Dis. 2010;4:e878. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 46] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 99. | Quan PL, Firth C, Street C, Henriquez JA, Petrosov A, Tashmukhamedova A, Hutchison SK, Egholm M, Osinubi MO, Niezgoda M. Identification of a severe acute respiratory syndrome coronavirus-like virus in a leaf-nosed bat in Nigeria. MBio. 2010;1:pii: e00208-10. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 103] [Cited by in F6Publishing: 122] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 100. | Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2013;41:D36-D42. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1924] [Cited by in F6Publishing: 1941] [Article Influence: 161.8] [Reference Citation Analysis (0)] |
| 101. | Pop M, Salzberg SL. Bioinformatics challenges of new sequencing technology. Trends Genet. 2008;24:142-149. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 320] [Cited by in F6Publishing: 234] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 102. | Henson J, Tischler G, Ning Z. Next-generation sequencing and large genome assemblies. Pharmacogenomics. 2012;13:901-915. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 100] [Cited by in F6Publishing: 68] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 103. | Sharma D, Priyadarshini P, Vrati S. Unraveling the web of viroinformatics: computational tools and databases in virus research. J Virol. 2015;89:1489-1501. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 53] [Article Influence: 5.3] [Reference Citation Analysis (1)] |
| 104. | Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403-410. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57523] [Cited by in F6Publishing: 60009] [Article Influence: 1765.0] [Reference Citation Analysis (0)] |