Copyright
©2014 Baishideng Publishing Group Inc.
World J Med Genet. May 27, 2014; 4(2): 6-18
Published online May 27, 2014. doi: 10.5496/wjmg.v4.i2.6
Published online May 27, 2014. doi: 10.5496/wjmg.v4.i2.6
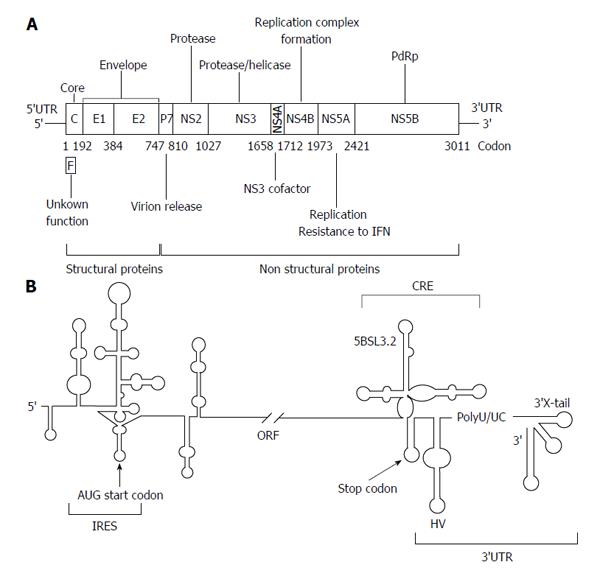
Figure 1 Genetic organization of the hepatitis C virus genomic RNA.
A: A schematic view of the hepatitis C virus (HCV) genome, showing the 5’ and 3’ untranslatable regions (UTRs) and the genes encoding for the different viral proteins. Numbers allude to codon positions; B: A detailed diagram of the secondary structure proposed for the 5’ and 3’ ends is pictured. The region required for internal ribosome entry site (IRES) activity is indicated. The 3’ end of the viral genomic RNA is organized into two well-defined structural elements: the cis-acting replicating element region containing the essential domain 5BSL3.2, and the 3’X-tail, separated by a hypervariable sequence (HV) and a polyU/UC tract. Start and stop translation codons, placed at positions 342 and 9371, respectively, are indicated by arrows. Numbers refer to the aminoacid positions according to the HCV Con1 isolate (GenBank accession number AJ238799). RdRp: RNA-dependent RNA polymerase; IFN: interferon; ORF: Open reading frame.
- Citation: Romero-López C, Berzal-Herranz A. Structure-function relationship in viral RNA genomes: The case of hepatitis C virus. World J Med Genet 2014; 4(2): 6-18
- URL: https://www.wjgnet.com/2220-3184/full/v4/i2/6.htm
- DOI: https://dx.doi.org/10.5496/wjmg.v4.i2.6