Published online Jan 19, 2024. doi: 10.5319/wjo.v11.i1.1
Peer-review started: September 18, 2023
First decision: December 12, 2023
Revised: December 21, 2023
Accepted: January 5, 2024
Article in press: January 5, 2024
Published online: January 19, 2024
Processing time: 120 Days and 21.5 Hours
Usher Syndrome (USH) is the most common deaf-blind syndrome, affecting approximately 1 in 6000 people in the deaf population. This genetic condition is characterized by a combination of hearing loss (HL), retinitis pigmentosa, and, in some cases, vestibular areflexia. Among the subtypes of USH, USH type 1 is considered the most severe form, presenting profound bilateral congenital deafness, vestibular areflexia, and early onset RP. USH type 2 is the most common form, exhibiting congenital moderate to severe HL for low frequencies and severe to profound HL for high frequencies. Conversely, type 3 is the rarest, initially manifesting mild symptoms during childhood that become more prominent in the first decades of life. The dual impact of USH on both visual and auditory senses significantly impairs patients’ quality of life, restricting their daily activities and interactions with society. To date, 9 genes have been confirmed so far for USH: MYO7A, USH1C, CDH23, PCDH15, USH1G, USH2A, ADGRV1, WHRN and CLRN1. These genes are inherited in an autosomal recessive manner and encode proteins expressed in the inner ear and retina, leading to functional loss. Although non-genetic methods can assist in patient triage and disease extension evaluation, genetic and molecular tests play a pivotal role in providing genetic counseling, enabling appropriate gene therapy, and facilitating timely cochlear implantation (CI). The CRISPR/Cas9 system and viral-based gene replacement therapy have recently emerged as highly promising techniques for treating USH. Regarding drug therapy, PTC-124 and Nb54 have been identified as promising drug interventions for genetic HL in USH. Simultaneously, CI has proven to be critical in the restoration of hearing. This review aims to summarize the genetic and molecular diagnosis of USH and highlight the importance of early diagnosis in guiding appropriate treatment strategies and improving patient prognosis.
Core Tip: Usher syndrome (USH) is a genetically inherited condition characterized by both hearing loss and vision impairment, leading to restrictions in daily activities and social interactions. Therefore, gaining a comprehensive understanding of the genetic and molecular aspects of this syndrome is of utmost importance to facilitate early intervention and enhance the quality of life for affected individuals. This review aims to summarize the genetic and molecular diagnosis and current treatments of USH and underscore the significance of early diagnosis in guiding appropriate treatment approaches.
- Citation: Cuzzuol BR, Apolonio JS, da Silva Júnior RT, de Carvalho LS, Santos LKS, Malheiro LH, Silva Luz M, Calmon MS, Crivellaro HL, Lemos FFB, Freire de Melo F. Usher syndrome: Genetic diagnosis and current therapeutic approaches. World J Otorhinolaryngol 2024; 11(1): 1-17
- URL: https://www.wjgnet.com/2218-6247/full/v11/i1/1.htm
- DOI: https://dx.doi.org/10.5319/wjo.v11.i1.1
Usher syndrome (USH) is the most common deaf-blind syndrome, affecting approximately 1 in 6000 people in the deaf population (approximately 400000 people worldwide)[1,2]. In the United States, this condition affects 16000 to 20000 people and represents 3% to 6% of early childhood deafness[3].
This genetic condition involves a combination of hearing loss (HL), visual impatience due to retinitis pigmentosa (RP), and, in some instances, vestibular areflexia leading to balance issues[4,5]. USH impact on ciliary cell functions classifies it as a ciliopathy, making it a complex disorder with significant consequences for affected individuals[6,7].
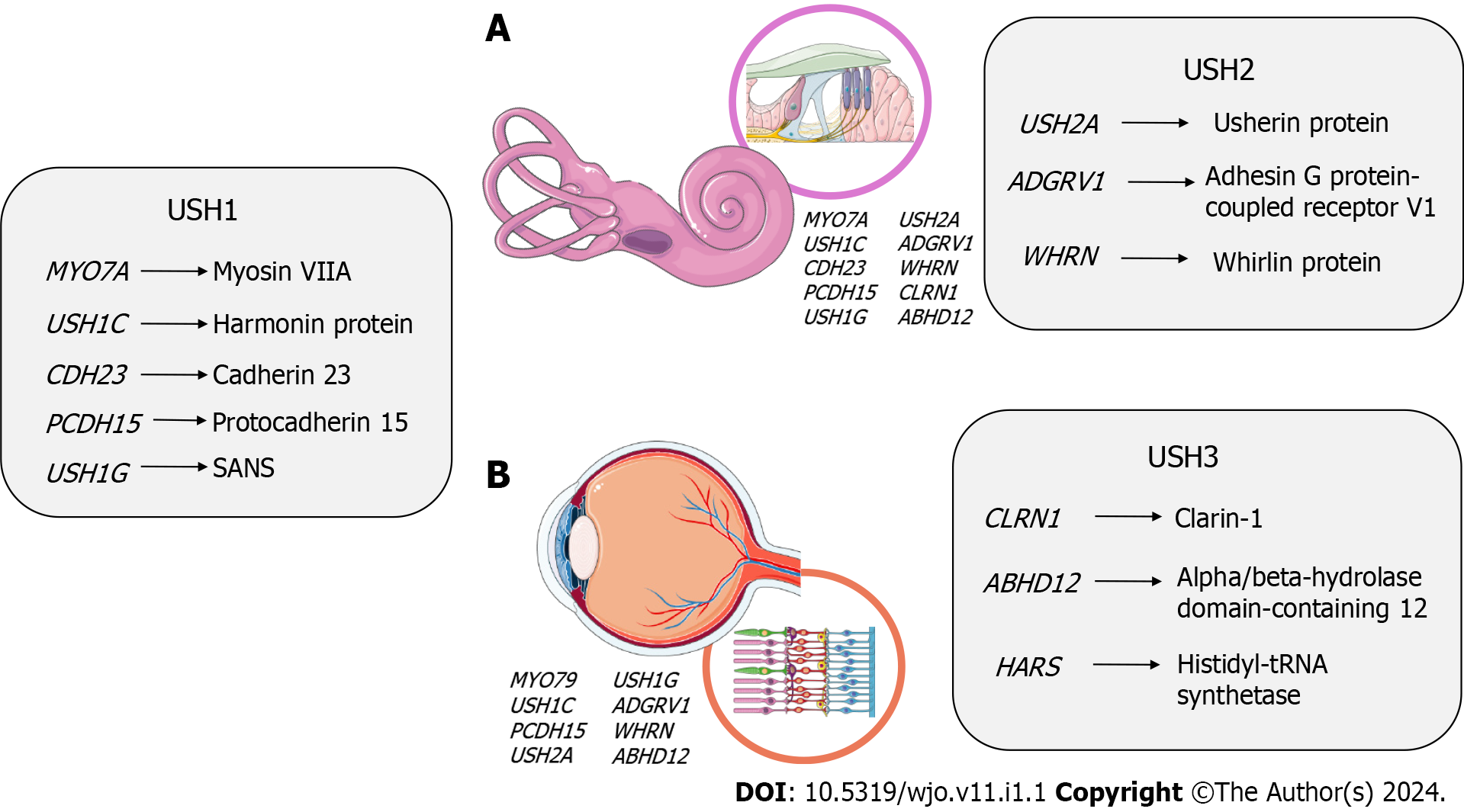
USH follows an autosomal recessive pattern, with nearly complete penetrance-about 100%. Currently, nine causative genes have been identified and confirmed for specific USH subtypes: MYO7A, USH1C, CDH23, PCDH15, and SANS for Usher type 1 (USH1); USH2A, ADGRV1, and WHRN for Usher type 2 (USH2); CLRN1 for Usher type 3 (USH3)[8].
The subtypes of USH are graded based on symptom severity and age of onset, with USH1 being the most severe form[3,9]. Patients do not recover auditory function or develop language ability unless fitted with a cochlear implantation (CI)[3,10]. CI not only rehabilitate hearing but also enhance speech intelligibility and significantly improve the overall quality of life[5].
USH significantly affects the quality of life of patients due to its dual impact on vision and hearing. Visual and hearing impairments can lead to limitations in daily activities and interaction with society, including social life, communication, access to information, and mobility[11,12]. As a result, these individuals often rely on other senses to communicate and to interact with society, such as tactile, kinesthetic, smell, haptic, and taste[13]. The progressive loss of vision necessitates constant adaptation in communication methods, which can be time-cognitive-and energy consuming. Vision loss also impedes the use of visual sign language[14,15]. Moreover, USH1 patients cannot rely on this form of communication due to the early onset of RP and vision loss, creating additional communication challenges[2,15,16]. Regarding vestibular dysfunction, early motor training can be applied to improve balance and mobility, enhancing proprioception and vision utilization[13].
At this moment, there is no effective treatment for USH[17]. Patients rely on supportive techniques like hearing aids and CI, that play a vital role in prolonging hearing time and restoring auditory function, but do not promote complete rehabilitation or inhibit the disease progression[18-20]. However, gene therapy has been making promising developments for visual and hearing restoration through CRISPR/Cas9 system, antisense oligonucleotides (ASOs) and viral vectors, like lentivirus and adenovirus, and drug therapy, with short interfering RNA (siRNA) and translational readthrough inducing drugs[14,15,16,21]. Early diagnosis appears to be crucial for enhancing the prognosis of USH patients, enabling timely intervention in young ages by promoting early delivery of gene therapy and rehabilitation of hearing, visual, and vestibular function in early childhood[2,24]. Thus, this review aims to summarize the genetic and molecular diagnosis of USH and correlate an early diagnosis with appropriate treatment and improved prognosis.
USH exhibits considerable clinical heterogeneity attributed to the characteristic genetic polymorphism of the disease. Variability is observed not only in symptomatology but also in disease intensity and aggressiveness among different USH subtypes. Particularly, HL emerges as a significant point of variation in these subtypes. USH1 is considered the most severe form of the disease, presenting with profound bilateral congenital deafness and vestibular areflexia, which remains non-progressive. USH1 is typically detected in the early years due to its impact on infants’ development, affecting language, balance, and motor skills[25]. It is essential to highlight that in the early years, vision compensates for deafness. However, as blindness emerges and progresses, this compensation becomes inadequate, leading to increased complexity in the condition[8].
The most prevalent form of USH is type 2, which is characterized by congenital bilateral HL of moderate to severe intensity[26]. The HL exhibits a characteristic sloping pattern, wherein low frequencies show mild to moderate impairment, while high frequencies experience severe to profound impairment. Diagnosis typically occurs around the age of 9[26], and the HL follows a non-progressive course, reaching stability around the third decade of life[13]. There is evidence suggesting a clear indication of HL progression, particularly in cases of USH2A[27]. Additionally, it is worth noting that vestibular issues are rare in these patients, as they typically maintain their balance within the normal range.
In turn, USH3 is the rarest form, initially showing mild affection during childhood, which becomes more prominent in the first decades of life and may vary until adulthood. Nevertheless, unlike USH2, USH3 presents with a progressive HL with a sloping pattern, leading to severe to profound deafness between the third and fourth decades of life[28]. Another distinguishing feature between USH2 and USH3 is vestibular alteration. USH3 exhibits variability in the expression of vestibular disorders due to the genetic variability in this subtype[28], which is not commonly observed in USH2, as it usually preserves vestibular function[29].
A common characteristic among all USH subtypes is that HL always occurs before visual changes, with the age of onset being the only variable. The visual impairment characteristic of USH is RP, a bilateral degeneration of the retina, starting peripherally and progressing towards the center. It initially affects the rods, leading to constriction of peripheral vision. As degeneration progresses, central blindness occurs, accompanied by impairment of the cones, which makes color perception and contrast sensitivity difficult[30]. RP varies among the subtypes; USH1 has an earlier onset, manifesting in the first decade of life and leading to complete blindness while still an infant. On the other hand, USH2 shows a later presentation of RP, and USH3 commonly exhibits RP during early youth[28]. In USH2, visual loss begins with mild changes, such as difficulty seeing in low light environments or night blindness, which can occur even in adulthood due to gene expression influences[27]. Fundus changes, such as atrophy or depigmentation of the retinal pigment epithelium, can also be observed[26].
Recent studies have attempted to investigate the correlation between USH and neurological and behavioral disorders, such as psychosis, schizophrenia, and autism in children with Usher[31]. These findings may have multifactorial origins due to the polygenic and pleiotropic nature of the disease. Additionally, they could contribute to the development of other alterations, as well as sensory and functional impairments, while lifestyle and behavior may also influence the development of clinical manifestations. Among all USH subtypes, some physical and psychological implications may be more pronounced in adults with USH, including fatigue, headaches, neck and shoulder pain, difficulty managing problems, and reduced social trust[13]. However, further research in this area is crucial to understand the correlation and impact of USH on the expression of other pathologies with neuropsychological impairment.
For a concise overview of the symptoms involved in each USH type, please refer to Table 1.
| Type | Hearing loss | Vision loss | Vestibular function |
| Type 1 | Profound bilateral hearing loss at birth[26,149] | Earlier onset, in the first decade of life[13,26] | Vestibular areflexia at birth[26] |
| Type 2 | Moderate to severe hearing loss at birth[13,24] | Late childhood or teens[26] | Normal[44] |
| Type 3 | Normal at birth, with progressive loss[28,164] | Commonly in early youth[13,26] | Variable[38] |
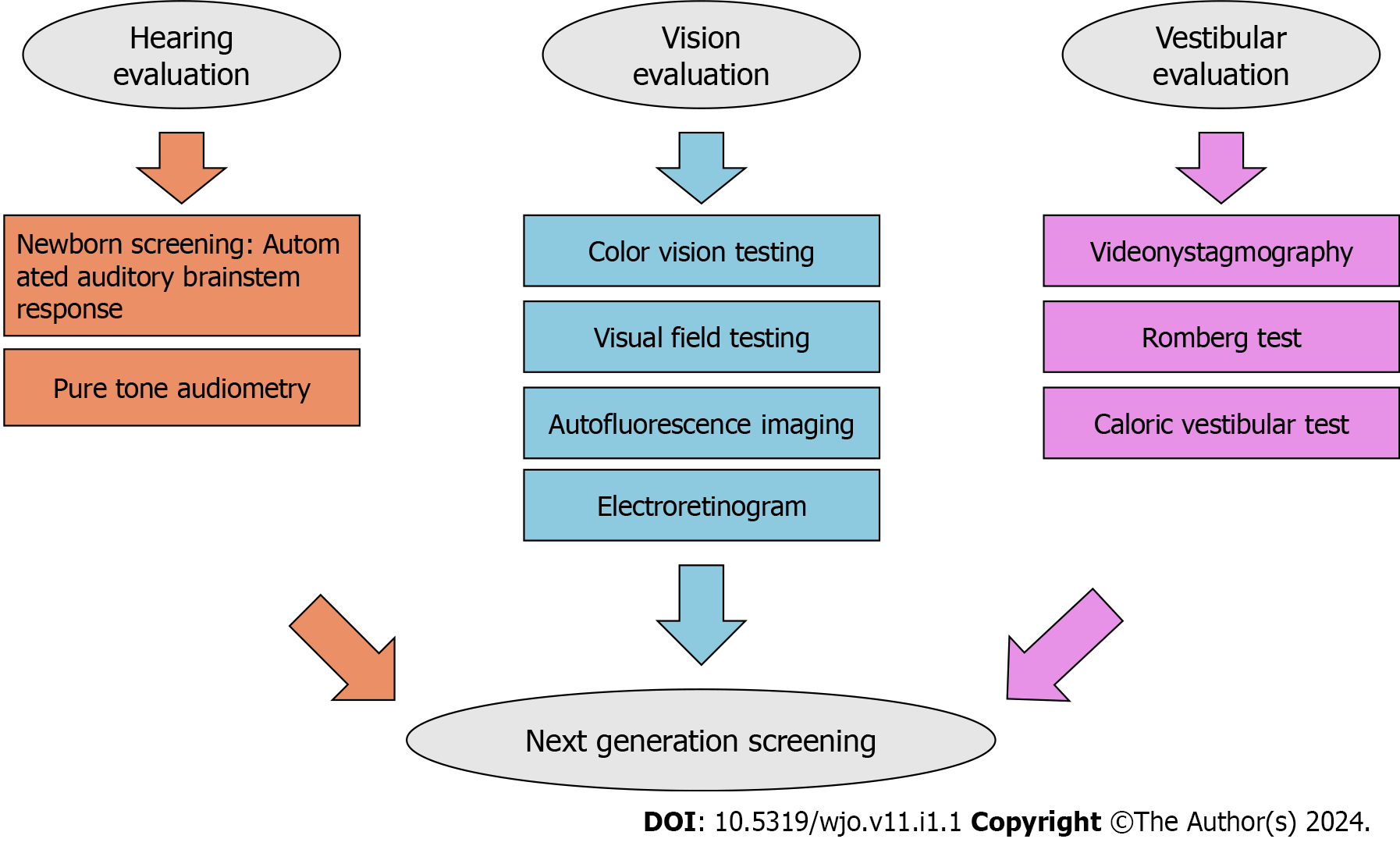
USH is classified into three subtypes based on the severity of symptoms, disease progression, and age of onset. Early diagnosis is crucial for effective clinical management and gaining a better understanding of genotype-phenotype relationships in this condition. It also helps anticipate therapeutic options, including CI[8,22]. Children with sensory impairments in vision and/or hearing have specific educational needs for their full development in different areas[32]. Non-genetic methods can aid in patient triage and evaluating the extent of the disease. Audiological tests, such as otoscopy, pure tone audiometry, and brainstem potential measurements (if specific frequencies are required), along with vestibular function tests-Romberg test, videonystagmography, and caloric vestibular test-posturography, and ophthalmological evaluation, such as color vision testing, visual field testing, autofluorescence imaging, and electroretinogram, can be utilized for this purpose[34]. In USH2, the most prevalent subtype, patients’ audiograms typically display a decreasing pattern with HL predominantly affecting elevated frequencies. Additionally, a reduced visual field creates a tunnel-like vision, and in severe cases, this field can be limited to 20 degrees or fewer. However, children with USH2A may exhibit unexpected HL in low frequencies and experience challenges in early vocabulary development[35]. Please refer to Figure 1 for a schematic of the diagnosis pathway.
The genetic landscape of clinical presentations in USH is notably complex, with approximately twelve genes linked to the three primary subtypes and other atypical phenotypes[32,35], and 9 genes have been confirmed to cause this condition so far. Among these genes, MYO7A, USH1C, CDH23, PCDH15, and USH1G are linked to USH1, while USH2A, ADGRV1, and WHRN are associated with USH2, and CLRN1 is linked to USH3[8]. These genes follow an autosomal recessive inheritance pattern and encode proteins expressed in the inner ear and retina[36]. Notably, the proteins encoded by these genes interact with each other, forming essential complexes crucial for the development and function of cilia in the inner ear and retina. When any of these proteins are absent or lose their function, it can lead to sensorineural degeneration in both the inner ear and retina[37].
Thus, prenatal diagnosis can become a useful tool, through the collection of amniotic fluid and genetic tests, especially when the clinical phenotype does not allow a clear clinical diagnosis[38]. Currently, the development of next generation sequencing (NGS) has greatly transformed the molecular diagnostics of both clinically and genetically highly heterogeneous neuro-sensory disorders, allowing rapid screening of large gene panels or the entire exome[33,39]. This technique has demonstrated to be time and cost-efficient: Genome can be sequenced within 3 d, at a cost of $1000 approximately. However, in cases of absence of insurance coverage or a public health system, patients might be unable to afford the tests[40,41].
USH differential diagnosis includes RP with non-genetic HL, simultaneous nonsyndromic RP and deafness, and “Usher-plus”. Regarding RP with non-genetic HL, HL can be explained by congenital infections, such as congenital cytomegalovirus, toxoplasmosis, and syphilis. Thus, conducting newborn tests for congenital infections is essential to distinguish between genetic and non-genetic HL[42,43]. Simultaneous nonsyndromic RP and deafness presents a clinical feature very similar to USH. In contrast to nonsyndromic RP, which exclusively affects the eyes, syndromic RP impacts additional neurosensory systems, including hearing. Hence, careful consideration of extraocular manifestations, family medical history, and genetic testing can help to elucidate the diagnosis[44-46]. Lastly, the presence of symptoms extending beyond deaf-blindness, termed “Usher-plus”, may indicate alternative genetic origins such as pseudo-USH, Heimler syndrome, cone-rod degeneration, retinal dystrophy, and premature aging syndrome. In such instances, NGS emerges as a valuable tool for distinguishing between these diverse diagnoses[45,46].
Moreover, establishing a precise molecular diagnosis is essential for genetic counseling and promoting appropriate gene therapy for affected individuals[32,35]. Table 2 summarizes the USH genes, comparing with the OMIM number, representative mutations, and respective geographic regions/ethnicities linked with these mutations.
| Genes | OMIM number | Representative mutations | Ethnic/geographic regions |
| MYO7A | 276903[47] | Deafness, autosomal dominant 11; Deafness, autosomal recessive 2; Usher syndrome, type 1B[47] | China, Japan, Pakistan, Netherlands, Iran, and Sweden[47] |
| USH1C | 605242[48] | Deafness, autosomal recessive 18A; Usher syndrome, type 1C[48] | Louisiana Acadians, and Lebanon[48] |
| CDH23 | 605516[49] | Deafness, autosomal recessive 12; Usher syndrome, type 1D[49] | Cuba, Germany, and Japan[49] |
| PCDH15 | 605514[50] | Deafness, autosomal recessive 23; Usher syndrome, type 1F[50] | Pakistan[50] |
| USH1G | 607696[51] | Usher syndrome, type 1G[51] | Tunisia, Germany, and Jordan[51] |
| USH2A | 608400[52] | Retinitis pigmentosa 39; Usher syndrome, type 2A[52] | Denmark, Norway, Spain, and Iraqi Jewish[52] |
| ADGRV1 | 602851[53] | Usher syndrome, type 2C; Usher syndrome, type 2C, GPR98/PDZD7 digenic; Familial Febrile Seizures 4[53] | Japan and France[53] |
| WHRN | 607928[54] | Deafness, autosomal recessive 31; Usher syndrome, type 2D[54] | Palestine, Tunisia, and Germany[54] |
| CLRN1 | 606397[55] | Retinitis pigmentosa 61; Usher syndrome, type 3A[55] | Italy, Ashkenazi Jewish population, and Spain[55] |
| ABHD121 | 613599[56] | Polyneuropathy, hearing loss, ataxia, retinitis pigmentosa, and cataract[56] | Norway, United Arab Emirates, United States, Algeria, Spain, and Netherlands[56] |
| HARS1 | 142810[57] | Charcot-Marie-Tooth disease, axonal, type 2W; Usher syndrome type 3B[57] | Old Order Amish families in Pennsylvania[57] |
In the following sections, we will provide detailed information about the specific genes associated with each clinical subtype of USH, summarized in Figure 2.
USH1 is the most severe subtype and is characterized by a severe to profound prelingual sensorineural HL (SNHL), early RP onset and vestibular alterations[7]. In addition to being clinically heterogeneous, USH is also genetically heterogeneous.
MYO7A: Consisting of 49 exons and encompassing an 87 kb genomic region, it was the first gene to be associated with USH[58,59]. MYO7A is the most prevalent gene among USH1, and pathogenic variants in this gene comprise 50% of the cases included in this subtype[60-62]. MYO7A gene encodes myosin VIIA, an actin-binding motor protein found in the cytoplasm and stereocilia of inner ear hair cells and within the connecting cilium and periciliary membranes of retinal photoreceptors[32,63]. Mutations in this gene are responsible for USH1B, affecting approximately 29% to 50% of cases[64-66]. In addition, they are also associated with non-syndromic recessive deafness 2, non-syndromic dominant deafness 11 and atypical USH[66]. This gene has been studied in gene therapy in phase I/II clinical trials[8].
USH1C: The genomic region of the USH1C gene extends to 51 kb and contains a total of 28 exons, eight of which having alternative splicing[67,68]. This gene has been described as causative of USH1 and non-syndromic HL[69]. Mutations in coding regions that are alternatively spliced (exons 15 and 22 to 28) have been related to non-syndromic HL[70] and mutations in constitutive coding to USH1. The USH1C gene encodes for the harmonin protein, a scaffold protein, expressed in several human cells, but with a key role in the USH protein network, especially in mechanosensitive hair cells of the inner ear and in photoreceptors and Müller glia cells[71]-consisting of 899 amino acids-, and a variety of scaffold proteins, all of which contain PDZ domains that are in charge of organizing protein complexes, and binding with USH1 and USH2 proteins in both the inner ear and retina[68,72,73]. Subcellular expression of harmonin is related to the USH1C phenotype[74].
CDH23:CDH23 is the second most frequently mutated gene in USH1 and has also been associated with non-syndromic autosomal recessive HL[75]. CDH23 gene encodes cadherin 23, a cell-cell adhesion protein with cadherin-like domains, important for the development and organization of stereocilia in the cochlea and vestibule, in addition to being part of the Usher protein network[13]. Nonsense, frameshift, splice-site and missense mutations in the CDH23 gene have been associated with USH1D. In addition, missense mutations that allow a residual protein function to be maintained, sufficient for the correct retina function, have been predominantly related to HL, whereas truncating variants are responsible for USH1[76-79].
PCDH15: The PCDH15 gene, which comprises a genomic region of 980 kb formed by 33 exons, is associated with both USH1 and non-syndromic HL, similar to CDH23[80-82]. PCDH15 gene encodes protocadherin 15, which belongs to the cadherin family, mediates calcium-dependent cell-cell adhesion and plays an important role in cochlear and retinal function. Variants of this gene are related to USH1F[83]. Furthermore, it has been reported that mutations in the PCDH15 and CDH23 genes can interact and lead to HL in digenic heterozygotes[84].
USH1G: Is the smallest USH gene, with a 7 kb genomic region and containing three exons, two of which are coding. It has been associated with USH1 and atypical USH, but, in recent years, it has also been described as possibly being responsible for non-syndromic HL[85-87]. This gene encodes a multivalent scaffold protein, SANS, which is associated with microtubule-based intracellular transport, regulation of endocytosis, and primary ciliogenesis. Deficiency in this protein and pathogenic USH1G mutations affect the splicing of genes related to cell proliferation and USH[88].
The diagnosis of a patient with USH2 initially involves a clinical evaluation. Congenital HL is bilateral and classified as moderate to severe. Before the age of 40, the acuity reduction tends to be stable, but it is more severe in high frequencies and milder in low frequencies. Vision loss can begin in early childhood and progress throughout life, showing no differences between the 3 types of USH[13]. Generally, equilibrium is not affected. The patient’s physical exam and general state of health typically do not present significant findings that contribute to a more definitive diagnosis. However, the medical history can reveal a recessive genetic inheritance pattern[30].
From the initial clinical suspicion, genetic panels can be requested to identify the specific genes related to USH2. Alternatively, if necessary, a comprehensive genetic sequencing can be performed, capable of encompassing both USH2 and other hereditary disorders that present similar symptoms, such as NGS. This approach allows for a more thorough investigation and a better chance of reaching a precise diagnosis[30]. The three genes more widely related to the USH2 are USH2A (USH Type 2A), ADGRV1 (adhesion G protein-coupled receptor V1) or GPR98 (G protein-coupled receptor 98), and WHRN (whirlin) or DFNB31 (Autosomal recessive non syndromic HL 31)[89,90].
USH2A:USH2A is more largely related to USH2. This gene is located in the long arm of chromosome 1, presenting two isoforms: a short, responsible for generating an extracellular protein present in the inner ear and retina, and a long, also dominant, related to the synthesis of usherin protein, which is expressed in the human retina, mainly in photoreceptor cells, and also contributes to development of cochlear hair cells[91,92]. Mutations in this gene are the most common cause of USH2 and are associated with moderate to severe congenital HL, photoreceptor degeneration, and non-syndromic autosomal recessive RP[93,94]. The impact of the clinical manifestations is extremely diverse, due to the different types of possible mutations[90]. Missense mutations in the USH2A gene can lead to the creation of proteins with altered structures due to the replacement of one amino acid with another, resulting in proteins that do not function properly or have reduced functionality. These types of mutations are often associated with later-onset symptoms. Mutations like nonsense, indel, or splicing mutations in the USH2A gene can indeed lead to early clinical manifestations in individuals due to the premature appearance of a stop codon and the synthesis of a smaller protein[95].
ADGRV1:ADGRV1 is located in the long arm of chromosome 5[96]. The ADGRV1 gene is accountable for the synthesis of the adhesion G protein-coupled receptor v1 (ADGRV1) protein, which is expressed in the retina, inner ear and central nervous system (CNS)[13,97]. The protein produced by ADGRV1 contributes to the development of hair cells of the inner ear, and the formation of the fibrous connections between the apical segment and cilia of photoreceptor cells, essential for proper signal transduction in both the auditory and visual systems[98]. Therefore, encoding disorders in this protein lead to impaired cell signaling and cell-cell adhesion, which may contribute to the progression of hearing and vision loss[97,99]. This gene can contribute to a higher incidence of HL, typically occurring within the first 10 years of life. Moreover, the HL related to the ADGRV1 mutations is often mild to severe, constant, and non-progressive, associated with physiologic maintenance of central vision and other structure and functional parameters until the end of the adult stage[96].
WHRN:WHRN is located on the long arm of chromosome 9[100]. WHRN encodes for the scaffold whirlin protein, which is expressed in the retina and inner ear[97]. It is involved in the formation of the periciliary membrane complex in photoreceptors and is essential for the development of hair cells in the cochlea and the elongation process of the stereocilia in hair cells[101,102]. Furthermore, WHRN plays an important role in supporting and stabilizing some protein interactions in the inner ear[103]. The potential implications of WHRN mutations are similar to those observed in other genes associated with this syndrome, such as USH2A, including retinal degeneration, HL and autosomal recessive non-syndromic deafness type 31 (DFNB31)[42,102,104]. The clinical manifestations vary depending on the protein region affected by the mutation, ranging from severe deafness associated with normal retinal function to moderate HL with progressive retinal degeneration[63].
USH3 is an autosomal recessive disorder that represents the rarest of the USH manifestations, with 1% to 6% of the total case[21,67]. The exceptions are Finnish and Ashkenazi Jewish families, in whom this percentage can reach 40% or more[8,32]. On the other hand, recent studies indicate that other locations may also have a higher prevalence, such as an epidemiologic clinical genetic study conducted in Japan in which USH3 accounted for 24.8%, the same percentage as USH1[105].
Another important aspect is the variable presentation of USH3 in relation to the other types, which is relevant for clinical diagnosis. SNHL is predominantly post lingual and progressive, more intense in the first and second decades of life, interspersed with periods of stability[8,106]. Its onset usually occurs in childhood but can manifest until around 35 years[32]. Even though patients may present with significant deafness, hearing is still preserved in the early stages of speech development, allowing speech not to be compromised[8,32,106]. As for RP, it is more likely to occur in the post-pubertal period, above 17-20 years, which starts with night blindness and progresses to visual field loss[8]. Finally, it is worth mentioning that half of the affected people will develop vestibular problems, unlike USH1 in which practically all patients present this deficit[8,106].
CLRN1: Regarding the genetic aspects, only the CLRN1 gene, which is located on chromosome 3q25, has been directly related to the development of USH3[13,106]. Thus, the literature classifies alterations in this gene as USH3A, with the possibility that, like the other types of USH, other variants will be described in the future[106]. This gene encodes a tetraspanin-like glycoprotein with 4 transmembrane domains called clarin-1, expressed in the hair bundle stereocilia present in the vestibular and cochlear systems, whose dysfunction interferes with the development of the inner ear neuroepithelium and can lead to deafness[8,32,106]. The exact mechanism that mutations in CLRN1 cause on the retina has not yet been fully clarified, with the hypothesis of its inappropriate expression directly in photoreceptor cells[32] or changes on the actin cytoskeleton of the cells of the Müller glia, preventing the processes of adhesion and transmembrane signaling[107]. Some alterations of this gene are common according to ethnicity, such as the nonsense mutation c.528T>G or p.Y176X in Finnish, the missense mutation c.144T>G or p.N48K in Ashkenazi Jewish families, or p.Y63X and p.C40G in Spanish, the latter being also related to early HL and speech problems, in addition to cases of areflexia[106].
ABHD12: Finally, there is the ABHD12 gene on the short arm of chromosome 20, which encodes the alpha/beta-hydrolase domain-containing 12, involved in the metabolism of endocannabinoid lipid transmitter 2-arachidonoyl glycerol[8,21]. Even though mutations in this gene are classically related to the development of polyneuropathy, HL, ataxia, RP, and cataracts (PHARC) syndrome, a recent study reported the occurrence of a nonsense variation of ABHD12 in two people from a Chinese family who were clinically diagnosed with USH3[108]. Thus, the authors raise the possibility that it can be related to the development of incomplete PHARC phenotypes or even interact with Usher proteins[21].
HARS: The HARS gene is located on chromosome 5q31 and encodes histidyl-tRNA synthetase, a cytoplasmic enzyme that enables the insertion of histidine into protein chains[8,13]. Nevertheless, there are just a few cases described in the literature that identify this mutation in patients diagnosed with USH3, which also needs to be better elucidated. In these individuals, in addition to the typical symptoms of USH3, neurological symptoms also have been reported[8,13,32] (Figure 2).
Gene therapy has shown promising advances in treating USH, offering the possibility of restoring hearing and vision. Viral-based gene replacement therapy and DNA editing techniques are currently at the forefront of research regarding hearing and vision recovery[2,22]. Gene therapy includes insertion, deletion, or replacement of whole genes, or the editing of pathogenic sequences with copies of the gene[2].
CRISPR/Cas9 system: Among these techniques, CRISPR/Cas9-a gene editing modality-stands out due to its high efficiency, simplicity, and targeting capability[23,109].
CRISPR/Cas9 system enables precise modifications to DNA sequences, making it a preferred choice for gene editing[5]. By creating double-strand breaks in DNA sequences, it allows for the modification and repair of a single base transition or transversion at the DNA level using a template DNA with the desired sequence[5,109]. Furthermore, the CRISPR/Cas9 system facilitates the mediation of large deletions and insertions, offering a more permanent treatment option[109]. While clinical application remains a challenge, gene therapy has shown successful results in animal models. The inner ear has proven to be an attractive target, as CRISPR/Cas9 has demonstrated success in a mouse model with autosomal dominant deafness[110,21]. In a pioneering study by Overlack et al[111], gene editing was attempted for USH using zinc finger nucleases to target the p.R31X mutation in the human USH1C gene. The in vitro results revealed partial repair of the USH1C gene and successful recovery of the harmonin protein in a p.R31X cell line following the transfection[5,113], indicating its potential on restoring function in inherited retinal disorders induced by disease-cause mutations. In terms of retinal interventions, mouse models of USH1C and USH1G have indicated that cones sensitive to oxidative stress may pose limitations in increasing the number of cone-like cells for RP treatment[112]. Consequently, identifying new suitable targets and optimizing efficiency remain significant challenges. A study employing viral delivery to fibroblasts derived from USH2 patients demonstrated a rescue of usherin expression. Despite these promising results, the efficiency was not significant enough to proceed to clinical trials[23]. Exon skipping is a mechanism that prompts the cellular machinery to bypass an exon containing a mutation. In the context of USH, exon skipping has proven to be a significant approach for mutations within exon 13, recurrent in USH2A[19]. In this way, the CRISPR/Cas9 system effectively excised the full-length exon containing the c.2299delG, c.2276G>T, and c.2802T>G mutations within exon 13 without inducing a frameshift. Moreover, the EDIT-101 trial (NCT03872479) demonstrated sufficient therapeutic safety by using dual CRISPR/Cas9 guides to target an intronic mutation, thereby reducing the risk of coding sequence break, and thus, of adverse effects and treatment failure[21]. As a result, the trial was able to progress through clinical trials for Leber Congenital Amaurosis treatment. Base editors, including cytosine and adenine base editors, have shown potential in repairing up to 30% of human pathogenic mutations, including USH2A gene[114,115]. USH2A can also be corrected through the replacement of the mutated codon with a conservative amino acid change[21].
Prime editing tools: Compared to CRISPR/Cas9, prime editing has a wider applicability, due to its capacity to mediate all base transitions in addition to small deletions up to 80 base pairs and insertions of up to 44 base pairs. In USH2A, prime editing tools enable the correction of all pathogenic point mutation in USH2A gene, such as c.2276G>T, 2802T>G, c.11864G>A, c.1256G>T, c.11156G>A, c.7595-2144A>G, c.8559-A>G, and c.9799T>C[21]. In turn, the study using CGF166 in patients with unilateral or bilateral severe-to-profound HL was pioneering in targeting the inner ear to proceed to clinical trials. It employed a recombinant adenovirus 5 (Ad5) vector containing cDNA encoding ATOH1, delivered via intra-labyrinthine infusion[116]. This study aimed to evaluate the safety of this method and its effects on pure tone audiometry over a two-year period. However, the trial was suspended due to lack of efficacy[2,116].
A major challenge faced by gene therapy in USH is that human hair cells are fully mature with response to sound at 25-27 wk of gestation, with damages in hair bundles and ribbon synapse already established[117,118]. In mice, however, the inner ear is still developing at birth, which explains why interventions can be successful in mouse models and may not have similar results in humans[118].
ASOs and viral vectors: ASOs and viral vectors are both valuable tools in gene therapy research. ASOs consist of short sequences of RNA/DNA that can inhibit translation by binding to complementary RNA strands[119]. In mouse models of USH1C, studies have demonstrated successful auditory and vestibular rescue by using ASOs to prevent the transcription of the c.216G > mutation[118,120,121,123].
Viral vectors, including adenovirus, adeno-associated virus (AAV), retrovirus, herpes simplex virus, and lentivirus, have also been utilized in inner ear therapies[122,124]. Among these, viral-based gene replacement therapy has shown superior efficiency compared to non-viral vectors, making it the most promising approach in gene therapy for USH at present[124]. Recombinant viral vectors possess inherent capabilities for cellular transduction, making them ideal for in vivo gene delivery. However, they do encounter challenges such as limited packaging capacities and potential immunogenicity[5]. AAV have emerged as the vector of choice for gene therapy due to their low immunogenicity, high cellular specificity, and sustained gene expression[5]. Nonetheless, their relatively small packaging capacity poses a challenge for treating USH, as the most common USH genes exceed the 4.7 kb packaging limit of AAV, such as the USH2A gene sequence[5,19,125]. To address this limitation, researchers have explored oversized AAV transgene constructs (fAAV) to increase AAV packaging capacity. A study by Allocca et al[126] identified rAAV2/5 as a potential packager capable of carrying up to 9.8 kb of genetic material in mouse models. Despite these efforts, some studies have indicated that fAAV vectors contain heterogeneous mixtures of gene fragments and do not carry full-length genes, reducing their efficiency in transduction capacity. Consequently, AAV vectors containing more than 5 kb of genetic material are less efficient in terms of transduction capacity[5]. Notwithstanding this limitation, AAV has shown potential in treating Usher retinal degeneration by effectively transducing photoreceptors[127]. On the other hand, adenovirus, another commonly used vector with a large packaging capacity, faces challenges due to its immunogenic response in most individuals, limiting its application in gene therapy[128]. Moreover, adenovirus exhibits low tropism for photoreceptor cells, further restricting its use in eye-targeted studies[129]. Lentiviruses, derived from the human immunodeficiency virus and possessing large packaging capacity, offer promise in USH research. In an in vivo study, lentiviral gene delivery corrected melanosome mislocalization and opsin accumulation at the photoreceptor connecting cilium in a mouse model of USH1B[130]. Additionally, subretinal delivery of UshStat to shake1 mice protected photoreceptors from light-induced degeneration, potentially aiding in preventing RP in USH1 patients[131].
Furthermore, Emptoz et al[132] achieved improvements in hearing and hair cell organization in USH1G mice by delivering rAAV2/8 containing USH1G cDNA to the inner ear. Similarly, Zou et al[133] demonstrated protein restoration of durable inner ear expression of Usher genes, and Pan et al[3] achieved gene and protein recovery of harmonin in a mouse model of USH1C using rAAV2/Anc80L65, a synthetically-produced AAV. AAV9-PHP.B has also been employed to package the Clrn-1 gene, providing rescue of low-frequency hearing (4-8 kHz) in a mouse model of USH3A deafness. However, the tropism and potential toxicity of AAV9-PHP.B in the CNS require further investigation[5,134].
Another promising approach for treating USH involves the ProQR commercial project, which demonstrated the retention of usherin function after ASO-mediated skipping of exon 13[113]. Gene replacement therapies like the ProQT trial for USH2A and UshStat for USH1 have shown progress, but they are limited in their ability to target specific mutations and provide only temporary effects[21]. UshStat, intended for USH1 patients, delivers MYO7A cDNA using a lentiviral vector, but its development did not progress further. On the other hand, the ProQR trial utilized ASOs (QR-421a) to bind to an mRNA splice site in the USH2A gene, leading to exon 13 skipping, and the trial advanced to phase 1/2. However, this treatment approach targets only mutations within exon 13, while other exons are skipped, resulting in deleterious frameshifts and a temporary treatment effect[19,21]. To address the challenge of targeting larger genes (> 4.7 kb), researchers have explored dual vector approaches. Nonetheless, this method also compromises efficiency, as demonstrated by Trapani et al[135], who achieved only 6% retinal transduction using dual AAV8 vectors. Dual vector approaches have been successful for the MYO7A gene in USH1B but present challenges for USH2A due to the gene’s large size[19]. Nonetheless, these approaches can be adapted using CRISPR tools for split delivery[5,19]. In terms of clinical applicability, these tools are still not ideal for therapeutic use, due to its struggle to achieve therapeutic efficiency, risk of immunogenicity, and safety concerns: CRIPR/Cas9 can induce double-strand breaks and lead to indel events and catastrophic chromothirpsis. On the other hand, FDA recently approved AAV for the treatment of adults with severe hemophilia A, at an acquisition cost of $2.9 million, considering it relatively safe[19,21].
RNA-based drug therapy offers a promising approach to silence or interfere with gain-of-function mutations and has been extensively studied for non-Usher genetic HL[4].
siRNA: siRNA has shown success in targeting the R75W allele variant in the GJB2 gene, which is associated with autosomal recessive HL. In the study by Maeda et al[136], siRNA treatment effectively suppressed GJB2 expression by more than 70%, preventing HL. Similarly, in the study by Shibata et al[137], microRNA was identified as a potential target for non-syndromic deafness caused by gain-of-function mutations in the TMC1 gene. ASOs provide a means to modulate gene expression by complementary binding to mRNA, making them useful for targeting pathogenic mutations[138,139]. One advantage of ASO gene delivery is that it is not constrained by gene size. However, the challenge lies in the relatively short half-life of ASOs, necessitating recurrent and invasive administration to the eye for a lasting effect when targeting retinal pathologies[19].
Translational readthrough inducing drugs: Translational Readthrough Inducing Drugs aim to prevent mRNA degradation by nonsense-mediated decay and avoid the production of truncated proteins caused by nonsense mutations[5,136,142]. This technique increases translational readthrough and is not gene or mutation-specific, while still allowing the cell to maintain expression regulation[5,141]. In the study by Nudelman et al[142], Nb30, a synthetic derivative of paromomycin, demonstrated potential in inducing nonsense suppression for USH1C mutations. Additionally, PTC-124, another translational readthrough inducer, showed superior recovery of protein function in USH1C mutations[143,144]. These findings, along with the study by Goldmann et al[145], identified PTC-124 and Nb54 as promising drug interventions for genetic HL in USH. In this view, PTC-124 is currently commercialized as Translarna for Duchenne Muscular Dystrophy, for the price of $3000 per gram of drug, which might be an obstacle for its acquisition[146]. Moreover, another limitation faced by drug therapy is the potential of these aminoglycosides-Nb30, PTC-124, and Nb54-to induce nephrotoxicity and ototoxicity, factor that may hinder its use in clinical practice[140,141].
CI is a widely used and effective treatment for restoring hearing and communication in patients with post-lingual severe-to-profound HL regarding their age, and in prelingually deaf infants[146,147]. This procedure utilizes a neuroprosthetic device placed within the cochlea, which allows direct interaction with the auditory nerve’s spiral ganglia, effectively bypassing earlier deficits in the hearing pathway[148].
USH1: Children with USH1 are the ideal candidates for CI, since they are considered prelingually deaf[149]. Early implantation, particularly before the age of two, is essential for better clinical outcomes and rehabilitation, improving communication skills[24,165]. In these patients, CI has shown to provide an almost identical phonological memory to normal-hearing children[151]. Similarly, Davies et al[149] systematic review revealed that patients implanted before two years old exhibited better vocabulary and speech perception, which can be attributed to their increased neural plasticity and continued auditory stimulation before experiencing substantial sensory deprivation. Thus, earlier CI is directly associated with better functional auditory outcomes and effective communication. Studies[151-153] have consistently shown that interventions before the age of 3.5 years can lead to a near-normal hearing in patients with congenital deafness. Chweya et al[153] systematic review of fifteen studies revealed that USH patients who received CI at younger ages showed improvements in pure-tone audiometry, speech perception, and expression outcomes after CI, while patients with prolonged auditory deprivation showed worse outcomes in sentence recognition and speech detection. Wu et al[154] systematic review also showed that earlier CI achieved better auditory outcomes than later implantation. However, delayed implantation leads to outcomes more in line with other deaf children with CI[149,151]. In addition, Remjasz-Jurek et al[155] evaluated auditory performance and speech intelligibility in children with USH up to 10 years after CI. The study found no statistically significant differences in auditory outcomes between USH and NS patients. Nonetheless, it also highlighted the superior results achieved by children implanted before the age of 3, compared to later implantation in USH and NS groups. Moreover, when implanted at later ages, patients often remain with severe HL when compared to patients who underwent CI as children, achieving worse results in terms of equivalent HL in decibel hearing level. A study[156] investigating outcomes of CI in both USH1 adults and children revealed that USH1 adults who underwent CI showed an average equivalent HL of 107.1 dB HL, whereas USH1 children had an equivalent of 84.4 dB HL. On the other hand, USH1 patients who did not receive CI remained profoundly deaf, with pure-tone average scores above 110 dB at 0.5 kHz, 1.0 kHz, and 2.0 kHz.
Recently, FDA approved CI for infants 9 mo of age or older with bilateral profound SNHL[153]. Considering that absence of auditory stimulation and auditory deprivation can cause degeneration across the central auditory system and provoke abnormal maturation in the auditory cortex, CI in the early months of life is decisive for adequate central auditory development[157]. Thus, the shorter the auditory deprivation, the more effective the auditory rehabilitation and language development after CI in infants. As Colletti et al[158] reported, CI before 12 mo of age resulted in near-normal vocabulary development compared to normal hearing children after ten years post-CI. CI between 12-24 mo showed a slight decrease in vocabulary development, while CI at 24-36 mo exhibited the most significant gap compared to normal hearing. Despite the benefits of CI in infants, it is necessary to consider its surgical risks. Infants are more vulnerable to hypothermia, airway collapse, fluid imbalance, and hemodynamic instability, as well as respiratory complications and other surgical complications, such as skin erythema, flap necrosis, and device migration[153]. Due to an underdeveloped mastoid tip anatomy, infants are also more vulnerable to facial nerve injury[153]. Nevertheless, early intervention is not always possible for all USH1 patients. Delayed diagnosis, socioeconomic factors, and lack of information about the disease can impede timely intervention, leading to difficulties in hearing, speaking, and communication when compared to those who received early CI implantation[149]. Moreover, CI is not commonly recommended for prelingual profoundly deaf adults, as they generally achieve lower auditory performance compared to post-lingual CI users[146]. Late CI also is not commonly recommended for non-progressive congenital HL[159]. Nonetheless, CI in prelingual adults still offers improvements in auditory scores, quality of life, and self-esteem[160-162]. Lahlou et al[147] retrospective study found that 44% of profound prelingually deaf adults patients showed increased auditory performance, with a mean speech intelligibility in silence of 65.0% ± 4.1%, similar to post-lingually profoundly deaf adults (67% to 76%) after CI. These results and improvements on speech recognition depend directly on previous hearing stimulations, development of hearing skills, and maturation of the auditory pathways. In cases when recovery of auditory response is not expected, CI can still benefits patients implanted in later ages by promoting access to sound, An observational study[163] involving 10 patients with USH1, implanted on average at 18.5 years, showed that only three patients improved their sentence recognition, but sound detection was achieved by most of the patients, which implies that late implantation can still provide access to sound. These results highlight the significance of CI in improving the quality of life of USH1 patients, though it may not yield optimal results in terms of speech understanding[148,149]. Thus, genetic testing and screenings in newborns are essential for early diagnosis and intervention in HL cases[149], as early identification allows timely CI, leading to improved outcomes and better quality of life[146,149].
USH2: In USH2, HL is progressive. For these patients, auditory rehabilitation typically begins with hearing aids in early childhood[24]. However, when severe HL, poor speech discrimination, and communication difficulties persist despite hearing aids, CI emerges as a crucial step in auditory rehabilitation[24,146]. Studies have consistently demonstrated significant improvements in pure tone audiometry, speech perception, and overall quality of life after CI, regardless of age at implantation. Hearing level had improved to 34 dB ± 9 dB, and word recognition had also better outcomes[24,164]. A retrospective case-control by Hartel et al[24] evaluated the performance of post lingually deaf adults with USH2A, with a mean age of implantation of 59 years. The results showed improvement in phoneme scores from 41% to 87% after CI. Thus, even when the mean age of implantation is relatively high, CI still results in notable improvements in hearing restoration. There were no differences in benefit between USH2A patients and control patients.
USH3: USH3 patients often experience HL before the third decade of life, either pre-lingual or post-lingual[164,165]. For these cases as well, CI presents a viable option for restoring hearing and improving communication[164]. For patients with residual hearing and progressive USH, the most beneficial timing for CI remains uncertain. Moreover, as USH often has a late diagnosis, CI commonly does not occur early, with an average age of 5.4 years, which impacts the effectiveness of rehabilitation efforts[166].
Reconsidering surgical indication might be necessary in patients with lack of auditory stimulations and relied solely on sign language. For these patients, CI may not attain measurable benefits[163]. Therefore, a comprehensive and multidisciplinary approach is essential for effective USH treatments, and must include psychologists, school educators, and various physicians, collaborating to address the diverse needs of all patients[146].
In conclusion, USH is a significant deaf-blind syndrome, affecting a considerable number of individuals worldwide. Although non-genetic methods can assist in patients’ triage, genetic testing is crucial for early diagnosis and effective clinical management, improving prognosis. Gene therapy has shown potential for treating USH, but faces challenges to produce significant and long-lasting therapeutic effects and guarantee safety. Thus, searching for smaller pieces of Cas proteins or prime editors and attempting to truncate existing proteins are possible strategies to bypass the packaging limit of AAV and other tools. Nonetheless, further investigations and clinical trials are necessary to validate the current approaches and bring them closer to clinical application. Another concern that should be addressed is the potential cost of these drugs and therapeutics and if it would be accessible to the population. Nevertheless, early diagnosis and intervention are essential in optimizing CI success, since better outcomes are directly associated with CI at younger ages. Moreover, early diagnosis may also enhance, in the future, the successful delivery of gene therapy for the inner ear and retina, improving auditory outcomes, language development, communication, and quality of life.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Otorhinolaryngology
Country/Territory of origin: Brazil
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Malik S, Pakistan S-Editor: Chen YL L-Editor: A P-Editor: Zhao S
| 1. | Nolen RM, Hufnagel RB, Friedman TB, Turriff AE, Brewer CC, Zalewski CK, King KA, Wafa TT, Griffith AJ, Brooks BP, Zein WM. Atypical and ultra-rare Usher syndrome: a review. Ophthalmic Genet. 2020;41:401-412. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in RCA: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 2. | de Joya EM, Colbert BM, Tang PC, Lam BL, Yang J, Blanton SH, Dykxhoorn DM, Liu X. Usher Syndrome in the Inner Ear: Etiologies and Advances in Gene Therapy. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in RCA: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 3. | Pan B, Askew C, Galvin A, Heman-Ackah S, Asai Y, Indzhykulian AA, Jodelka FM, Hastings ML, Lentz JJ, Vandenberghe LH, Holt JR, Géléoc GS. Gene therapy restores auditory and vestibular function in a mouse model of Usher syndrome type 1c. Nat Biotechnol. 2017;35:264-272. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited in This Article: ] [Cited by in Crossref: 187] [Cited by in RCA: 220] [Article Influence: 27.5] [Reference Citation Analysis (0)] |
| 4. | Géléoc GGS, El-Amraoui A. Disease mechanisms and gene therapy for Usher syndrome. Hear Res. 2020;394:107932. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in RCA: 49] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 5. | French LS, Mellough CB, Chen FK, Carvalho LS. A Review of Gene, Drug and Cell-Based Therapies for Usher Syndrome. Front Cell Neurosci. 2020;14:183. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 6. | Tsang SH, Aycinena ARP, Sharma T. Ciliopathy: Usher Syndrome. Adv Exp Med Biol. 2018;1085:167-170. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in RCA: 19] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 7. | Fuster-García C, García-Bohórquez B, Rodríguez-Muñoz A, Aller E, Jaijo T, Millán JM, García-García G. Usher Syndrome: Genetics of a Human Ciliopathy. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in RCA: 54] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 8. | Delmaghani S, El-Amraoui A. The genetic and phenotypic landscapes of Usher syndrome: from disease mechanisms to a new classification. Hum Genet. 2022;141:709-735. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in RCA: 53] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 9. | Reiners J, Nagel-Wolfrum K, Jürgens K, Märker T, Wolfrum U. Molecular basis of human Usher syndrome: deciphering the meshes of the Usher protein network provides insights into the pathomechanisms of the Usher disease. Exp Eye Res. 2006;83:97-119. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 178] [Cited by in RCA: 197] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 10. | Taiber S, Avraham KB. Genetic Therapies for Hearing Loss: Accomplishments and Remaining Challenges. Neurosci Lett. 2019;713:134527. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in RCA: 11] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 11. | Ehn M, Wahlqvist M, Danermark B, Dahlström Ö, Möller C. Health, work, social trust, and financial situation in persons with Usher syndrome type 1. Work. 2018;60:209-220. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in RCA: 9] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 12. | Ehn M, Wahlqvist M, Möller C, Anderzén-Carlsson A. The lived experiences of work and health of people living with deaf-blindness due to Usher syndrome type 2. Int J Qual Stud Health Well-being. 2020;15:1846671. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited in This Article: ] [Reference Citation Analysis (0)] |
| 13. | Castiglione A, Möller C. Usher Syndrome. Audiol Res. 2022;12:42-65. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in RCA: 45] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 14. | Ehn M, Anderzén-Carlsson A, Möller C, Wahlqvist M. Life strategies of people with deafblindness due to Usher syndrome type 2a - a qualitative study. Int J Qual Stud Health Well-being. 2019;14:1656790. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in RCA: 6] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 15. | Wahlqvist M, Möller C, Möller K, Danermark B. Similarities and Differences in Health, Social Trust, and Financial Situation in People With Usher Syndrome, a Bio-Psychosocial Perspective. Front Psychol. 2020;11:1760. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in RCA: 6] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 16. | Bonnet C, Grati M, Marlin S, Levilliers J, Hardelin JP, Parodi M, Niasme-Grare M, Zelenika D, Délépine M, Feldmann D, Jonard L, El-Amraoui A, Weil D, Delobel B, Vincent C, Dollfus H, Eliot MM, David A, Calais C, Vigneron J, Montaut-Verient B, Bonneau D, Dubin J, Thauvin C, Duvillard A, Francannet C, Mom T, Lacombe D, Duriez F, Drouin-Garraud V, Thuillier-Obstoy MF, Sigaudy S, Frances AM, Collignon P, Challe G, Couderc R, Lathrop M, Sahel JA, Weissenbach J, Petit C, Denoyelle F. Complete exon sequencing of all known Usher syndrome genes greatly improves molecular diagnosis. Orphanet J Rare Dis. 2011;6:21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited in This Article: ] [Cited by in Crossref: 81] [Cited by in RCA: 81] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 17. | Arcous M, Putois O, Dalle-Nazébi S, Kerbourch S, Cariou A, Ben Aissa I, Marlin S, Potier R. Psychosocial determinants associated with quality of life in people with usher syndrome. A scoping review. Disabil Rehabil. 2020;42:2809-2820. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 18. | El-Amraoui A, Petit C. The retinal phenotype of Usher syndrome: pathophysiological insights from animal models. C R Biol. 2014;337:167-177. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 33] [Cited by in RCA: 39] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 19. | Major L, McClements ME, MacLaren RE. New CRISPR Tools to Correct Pathogenic Mutations in Usher Syndrome. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited in This Article: ] [Reference Citation Analysis (0)] |
| 20. | Young NM, Johnson JC, Mets MB, Hain TC. Cochlear implants in young children with Usher's syndrome. Ann Otol Rhinol Laryngol Suppl. 1995;166:342-345. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in RCA: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 21. | Major L, McClements ME, MacLaren RE. A Review of CRISPR Tools for Treating Usher Syndrome: Applicability, Safety, Efficiency, and In Vivo Delivery. Int J Mol Sci. 2023;24. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Reference Citation Analysis (0)] |
| 22. | Nisenbaum E, Prentiss S, Yan D, Nourbakhsh A, Smeal M, Holcomb M, Cejas I, Telischi F, Liu XZ. Screening Strategies for Deafness Genes and Functional Outcomes in Cochlear Implant Patients. Otol Neurotol. 2021;42:180-187. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in RCA: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 23. | Fukui H, Raphael Y. Gene therapy for the inner ear. Hear Res. 2013;297:99-105. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in RCA: 45] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 24. | Hartel BP, van Nierop JWI, Huinck WJ, Rotteveel LJC, Mylanus EAM, Snik AF, Kunst HPM, Pennings RJE. Cochlear Implantation in Patients With Usher Syndrome Type IIa Increases Performance and Quality of Life. Otol Neurotol. 2017;38:e120-e127. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in RCA: 24] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 25. | Toms M, Pagarkar W, Moosajee M. Usher syndrome: clinical features, molecular genetics and advancing therapeutics. Ther Adv Ophthalmol. 2020;12:2515841420952194. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in RCA: 44] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 26. | Blanco-Kelly F, Jaijo T, Aller E, Avila-Fernandez A, López-Molina MI, Giménez A, García-Sandoval B, Millán JM, Ayuso C. Clinical aspects of Usher syndrome and the USH2A gene in a cohort of 433 patients. JAMA Ophthalmol. 2015;133:157-164. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in RCA: 54] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 27. | Sadeghi M, Cohn ES, Kelly WJ, Kimberling WJ, Tranebjoerg L, Möller C. Audiological findings in Usher syndrome types IIa and II (non-IIa). Int J Audiol. 2004;43:136-143. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 27] [Cited by in RCA: 24] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 28. | Sadeghi M, Cohn ES, Kimberling WJ, Tranebjaerg L, Möller C. Audiological and vestibular features in affected subjects with USH3: a genotype/phenotype correlation. Int J Audiol. 2005;44:307-316. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in RCA: 22] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 29. | Geng R, Geller SF, Hayashi T, Ray CA, Reh TA, Bermingham-McDonogh O, Jones SM, Wright CG, Melki S, Imanishi Y, Palczewski K, Alagramam KN, Flannery JG. Usher syndrome IIIA gene clarin-1 is essential for hair cell function and associated neural activation. Hum Mol Genet. 2009;18:2748-2760. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 58] [Cited by in RCA: 66] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 30. | Koenekoop R, Arriaga M, Trzupek KM, Lentz J. Usher Syndrome Type II. 1999 Dec 10. In: GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 1993–. [PubMed] [Cited in This Article: ] |
| 31. | Domanico D, Fragiotta S, Cutini A, Grenga PL, Vingolo EM. Psychosis, Mood and Behavioral Disorders in Usher Syndrome: Review of the Literature. Med Hypothesis Discov Innov Ophthalmol. 2015;4:50-55. [PubMed] [Cited in This Article: ] |
| 32. | Nisenbaum E, Thielhelm TP, Nourbakhsh A, Yan D, Blanton SH, Shu Y, Koehler KR, El-Amraoui A, Chen Z, Lam BL, Liu X. Review of Genotype-Phenotype Correlations in Usher Syndrome. Ear Hear. 2022;43:1-8. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in RCA: 19] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 33. | Bahena P, Daftarian N, Maroofian R, Linares P, Villalobos D, Mirrahimi M, Rad A, Doll J, Hofrichter MAH, Koparir A, Röder T, Han S, Sabbaghi H, Ahmadieh H, Behboudi H, Villanueva-Mendoza C, Cortés-Gonzalez V, Zamora-Ortiz R, Kohl S, Kuehlewein L, Darvish H, Alehabib E, Arenas-Sordo ML, Suri F, Vona B, Haaf T. Unraveling the genetic complexities of combined retinal dystrophy and hearing impairment. Hum Genet. 2022;141:785-803. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in RCA: 7] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 34. | Seeliger MW, Fischer MD, Pfister M. [Usher syndrome: clinical features, diagnostic options, and therapeutic prospects]. Ophthalmologe. 2009;106:505-511. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 35. | Bonnet C, Riahi Z, Chantot-Bastaraud S, Smagghe L, Letexier M, Marcaillou C, Lefèvre GM, Hardelin JP, El-Amraoui A, Singh-Estivalet A, Mohand-Saïd S, Kohl S, Kurtenbach A, Sliesoraityte I, Zobor D, Gherbi S, Testa F, Simonelli F, Banfi S, Fakin A, Glavač D, Jarc-Vidmar M, Zupan A, Battelino S, Martorell Sampol L, Claveria MA, Catala Mora J, Dad S, Møller LB, Rodriguez Jorge J, Hawlina M, Auricchio A, Sahel JA, Marlin S, Zrenner E, Audo I, Petit C. An innovative strategy for the molecular diagnosis of Usher syndrome identifies causal biallelic mutations in 93% of European patients. Eur J Hum Genet. 2016;24:1730-1738. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited in This Article: ] [Cited by in Crossref: 61] [Cited by in RCA: 70] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 36. | Jouret G, Poirsier C, Spodenkiewicz M, Jaquin C, Gouy E, Arndt C, Labrousse M, Gaillard D, Doco-Fenzy M, Lebre AS. Genetics of Usher Syndrome: New Insights From a Meta-analysis. Otol Neurotol. 2019;40:121-129. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 42] [Cited by in RCA: 61] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 37. | Koenekoop RK, Arriaga MA, Trzupek KM, Lentz JJ. Usher Syndrome Type I. 1999 Dec 10. In: GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 1993–. [PubMed] [Cited in This Article: ] |
| 38. | Zhou C, Xiao Y, Xie H, Liu S, Wang J. A novel USH2A variant in a patient with hearing loss and prenatal diagnosis of a familial fetus: a case report. BMC Med Genomics. 2021;14:200. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited in This Article: ] [Cited by in Crossref: 1] [Reference Citation Analysis (0)] |
| 39. | Oishi M, Oishi A, Gotoh N, Ogino K, Higasa K, Iida K, Makiyama Y, Morooka S, Matsuda F, Yoshimura N. Comprehensive molecular diagnosis of a large cohort of Japanese retinitis pigmentosa and Usher syndrome patients by next-generation sequencing. Invest Ophthalmol Vis Sci. 2014;55:7369-7375. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 110] [Cited by in RCA: 114] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 40. | Kumar KR, Cowley MJ, Davis RL. Next-Generation Sequencing and Emerging Technologies. Semin Thromb Hemost. 2019;45:661-673. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 93] [Cited by in RCA: 148] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 41. | Johansen Taber KA, Dickinson BD, Wilson M. The promise and challenges of next-generation genome sequencing for clinical care. JAMA Intern Med. 2014;174:275-280. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 110] [Cited by in RCA: 110] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 42. | Kimberling WJ, Hildebrand MS, Shearer AE, Jensen ML, Halder JA, Trzupek K, Cohn ES, Weleber RG, Stone EM, Smith RJ. Frequency of Usher syndrome in two pediatric populations: Implications for genetic screening of deaf and hard of hearing children. Genet Med. 2010;12:512-516. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 152] [Cited by in RCA: 169] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 43. | Morton CC, Nance WE. Newborn hearing screening--a silent revolution. N Engl J Med. 2006;354:2151-2164. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 1024] [Cited by in RCA: 1121] [Article Influence: 59.0] [Reference Citation Analysis (0)] |
| 44. | Verbakel SK, van Huet RAC, Boon CJF, den Hollander AI, Collin RWJ, Klaver CCW, Hoyng CB, Roepman R, Klevering BJ. Non-syndromic retinitis pigmentosa. Prog Retin Eye Res. 2018;66:157-186. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 331] [Cited by in RCA: 580] [Article Influence: 82.9] [Reference Citation Analysis (0)] |
| 45. | Gerth-Kahlert C, Koller S. [Ciliopathies]. Klin Monbl Augenheilkd. 2018;235:264-272. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 46. | Boycott KM, Innes AM. When One Diagnosis Is Not Enough. N Engl J Med. 2017;376:83-85. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in RCA: 23] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 47. | Online Mendelian Inheritance in Man. OMIM®. Johns Hopkins University, Baltimore, MD. MIM Number: MYO7A. 2023. [cited 10 December 2023]. Available from: https://omim.org/entry/276903. [Cited in This Article: ] |
| 48. | Online Mendelian Inheritance in Man. OMIM®. Johns Hopkins University, Baltimore, MD. MIM Number: USH1C. 2023. [cited 10 December 2023]. Available from: https://omim.org/entry/605242. [Cited in This Article: ] |
| 49. | Online Mendelian Inheritance in Man. OMIM®. Johns Hopkins University, Baltimore, MD. MIM Number: CDH23. 2023. [cited 10 December 2023]. Available from: https://omim.org/entry/605516. [Cited in This Article: ] |
| 50. | Online Mendelian Inheritance in Man. OMIM®. Johns Hopkins University, Baltimore, MD. MIM Number: PCDH15. 2023. [cited 10 December 2023]. Available from: https://omim.org/entry/605514. [Cited in This Article: ] |
| 51. | Online Mendelian Inheritance in Man. OMIM®. Johns Hopkins University, Baltimore, MD. MIM Number: USH1G. 2023. [cited 10 December 2023]. Available from: https://omim.org/entry/607696. [Cited in This Article: ] |
| 52. | Online Mendelian Inheritance in Man. OMIM®. Johns Hopkins University, Baltimore, MD. MIM Number: USH2A. 2023. [cited 10 December 2023]. Available from: https://omim.org/entry/608400. [Cited in This Article: ] |
| 53. | Online Mendelian Inheritance in Man. OMIM®. Johns Hopkins University, Baltimore, MD. MIM Number: ADGRV1. 2023. [cited 10 December 2023]. Available from: https://omim.org/entry/602851. [Cited in This Article: ] |
| 54. | Online Mendelian Inheritance in Man. OMIM®. Johns Hopkins University, Baltimore, MD. MIM Number: WHRN. 2023. [cited 10 December 2023]. Available from: https://omim.org/entry/607928. [Cited in This Article: ] |
| 55. | Online Mendelian Inheritance in Man. OMIM®. Johns Hopkins University, Baltimore, MD. MIM Number: CLRN1. 2023. [cited 10 December 2023]. Available from: https://omim.org/entry/606397. [Cited in This Article: ] |
| 56. | Online Mendelian Inheritance in Man. OMIM®. Johns Hopkins University, Baltimore, MD. MIM Number: ABHD12. 2023. [cited 10 December 2023]. Available from: https://omim.org/entry/613599. [Cited in This Article: ] |
| 57. | Online Mendelian Inheritance in Man. OMIM®. Johns Hopkins University, Baltimore, MD. MIM Number: HARS. 2023. [cited 10 December 2023]. Available from: https://omim.org/entry/142810. [Cited in This Article: ] |
| 58. | Kimberling WJ, Möller CG, Davenport S, Priluck IA, Beighton PH, Greenberg J, Reardon W, Weston MD, Kenyon JB, Grunkemeyer JA. Linkage of Usher syndrome type I gene (USH1B) to the long arm of chromosome 11. Genomics. 1992;14:988-994. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 133] [Cited by in RCA: 136] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 59. | Weil D, Blanchard S, Kaplan J, Guilford P, Gibson F, Walsh J, Mburu P, Varela A, Levilliers J, Weston MD. Defective myosin VIIA gene responsible for Usher syndrome type 1B. Nature. 1995;374:60-61. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 766] [Cited by in RCA: 766] [Article Influence: 25.5] [Reference Citation Analysis (0)] |
| 60. | Jaijo T, Aller E, Beneyto M, Najera C, Graziano C, Turchetti D, Seri M, Ayuso C, Baiget M, Moreno F, Morera C, Perez-Garrigues H, Millan JM. MYO7A mutation screening in Usher syndrome type I patients from diverse origins. J Med Genet. 2007;44:e71. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in RCA: 42] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 61. | Fuster-García C, García-García G, Jaijo T, Fornés N, Ayuso C, Fernández-Burriel M, Sánchez-De la Morena A, Aller E, Millán JM. High-throughput sequencing for the molecular diagnosis of Usher syndrome reveals 42 novel mutations and consolidates CEP250 as Usher-like disease causative. Sci Rep. 2018;8:17113. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in RCA: 25] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 62. | Liu XZ, Hope C, Walsh J, Newton V, Ke XM, Liang CY, Xu LR, Zhou JM, Trump D, Steel KP, Bundey S, Brown SD. Mutations in the myosin VIIA gene cause a wide phenotypic spectrum, including atypical Usher syndrome. Am J Hum Genet. 1998;63:909-912. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 76] [Cited by in RCA: 87] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 63. | Mathur PD, Yang J. Usher syndrome and non-syndromic deafness: Functions of different whirlin isoforms in the cochlea, vestibular organs, and retina. Hear Res. 2019;375:14-24. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in RCA: 40] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 64. | Riazuddin S, Nazli S, Ahmed ZM, Yang Y, Zulfiqar F, Shaikh RS, Zafar AU, Khan SN, Sabar F, Javid FT, Wilcox ER, Tsilou E, Boger ET, Sellers JR, Belyantseva IA, Riazuddin S, Friedman TB. Mutation spectrum of MYO7A and evaluation of a novel nonsyndromic deafness DFNB2 allele with residual function. Hum Mutat. 2008;29:502-511. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 72] [Cited by in RCA: 79] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 65. | Subirà O, Català-Mora J, Díaz-Cascajosa J, Padrón-Pérez N, Claveria MA, Coll-Alsina N, Bonnet C, Petit C, Caminal JM, Prat J. Retinal findings in pediatric patients with Usher syndrome Type 1 due to mutations in MYO7A gene. Eye (Lond). 2020;34:499-506. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 2] [Cited by in RCA: 5] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 66. | Millán JM, Aller E, Jaijo T, Blanco-Kelly F, Gimenez-Pardo A, Ayuso C. An update on the genetics of usher syndrome. J Ophthalmol. 2011;2011:417217. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited in This Article: ] [Cited by in Crossref: 112] [Cited by in RCA: 135] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 67. | Mathur P, Yang J. Usher syndrome: Hearing loss, retinal degeneration and associated abnormalities. Biochim Biophys Acta. 2015;1852:406-420. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 191] [Cited by in RCA: 222] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 68. | Bitner-Glindzicz M, Lindley KJ, Rutland P, Blaydon D, Smith VV, Milla PJ, Hussain K, Furth-Lavi J, Cosgrove KE, Shepherd RM, Barnes PD, O'Brien RE, Farndon PA, Sowden J, Liu XZ, Scanlan MJ, Malcolm S, Dunne MJ, Aynsley-Green A, Glaser B. A recessive contiguous gene deletion causing infantile hyperinsulinism, enteropathy and deafness identifies the Usher type 1C gene. Nat Genet. 2000;26:56-60. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 221] [Cited by in RCA: 239] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 69. | Verpy E, Leibovici M, Zwaenepoel I, Liu XZ, Gal A, Salem N, Mansour A, Blanchard S, Kobayashi I, Keats BJ, Slim R, Petit C. A defect in harmonin, a PDZ domain-containing protein expressed in the inner ear sensory hair cells, underlies Usher syndrome type 1C. Nat Genet. 2000;26:51-55. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 332] [Cited by in RCA: 321] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 70. | Jain PK, Lalwani AK, Li XC, Singleton TL, Smith TN, Chen A, Deshmukh D, Verma IC, Smith RJ, Wilcox ER. A gene for recessive nonsyndromic sensorineural deafness (DFNB18) maps to the chromosomal region 11p14-p15.1 containing the Usher syndrome type 1C gene. Genomics. 1998;50:290-292. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in RCA: 35] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 71. | Reiners J, van Wijk E, Märker T, Zimmermann U, Jürgens K, te Brinke H, Overlack N, Roepman R, Knipper M, Kremer H, Wolfrum U. Scaffold protein harmonin (USH1C) provides molecular links between Usher syndrome type 1 and type 2. Hum Mol Genet. 2005;14:3933-3943. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 121] [Cited by in RCA: 128] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 72. | Schäfer J, Wenck N, Janik K, Linnert J, Stingl K, Kohl S, Nagel-Wolfrum K, Wolfrum U. The Usher syndrome 1C protein harmonin regulates canonical Wnt signaling. Front Cell Dev Biol. 2023;11:1130058. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Reference Citation Analysis (0)] |
| 73. | Sheng M, Sala C. PDZ domains and the organization of supramolecular complexes. Annu Rev Neurosci. 2001;24:1-29. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 923] [Cited by in RCA: 941] [Article Influence: 39.2] [Reference Citation Analysis (0)] |
| 74. | Donaldson TN, Jennings KT, Cherep LA, McNeela AM, Depreux FF, Jodelka FM, Hastings ML, Wallace DG. Antisense oligonucleotide therapy rescues disruptions in organization of exploratory movements associated with Usher syndrome type 1C in mice. Behav Brain Res. 2018;338:76-87. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in RCA: 13] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 75. | Nagel-Wolfrum K, Fadl BR, Becker MM, Wunderlich KA, Schäfer J, Sturm D, Fritze J, Gür B, Kaplan L, Andreani T, Goldmann T, Brooks M, Starostik MR, Lokhande A, Apel M, Fath KR, Stingl K, Kohl S, DeAngelis MM, Schlötzer-Schrehardt U, Kim IK, Owen LA, Vetter JM, Pfeiffer N, Andrade-Navarro MA, Grosche A, Swaroop A, Wolfrum U. Expression and subcellular localization of USH1C/harmonin in human retina provides insights into pathomechanisms and therapy. Hum Mol Genet. 2023;32:431-449. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 76. | Bolz H, von Brederlow B, Ramírez A, Bryda EC, Kutsche K, Nothwang HG, Seeliger M, del C-Salcedó Cabrera M, Vila MC, Molina OP, Gal A, Kubisch C. Mutation of CDH23, encoding a new member of the cadherin gene family, causes Usher syndrome type 1D. Nat Genet. 2001;27:108-112. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 320] [Cited by in RCA: 339] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 77. | Astuto LM, Bork JM, Weston MD, Askew JW, Fields RR, Orten DJ, Ohliger SJ, Riazuddin S, Morell RJ, Khan S, Kremer H, van Hauwe P, Moller CG, Cremers CW, Ayuso C, Heckenlively JR, Rohrschneider K, Spandau U, Greenberg J, Ramesar R, Reardon W, Bitoun P, Millan J, Legge R, Friedman TB, Kimberling WJ. CDH23 mutation and phenotype heterogeneity: a profile of 107 diverse families with Usher syndrome and nonsyndromic deafness. Am J Hum Genet. 2002;71:262-275. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 170] [Cited by in RCA: 171] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 78. | Baux D, Faugère V, Larrieu L, Le Guédard-Méreuze S, Hamroun D, Béroud C, Malcolm S, Claustres M, Roux AF. UMD-USHbases: a comprehensive set of databases to record and analyse pathogenic mutations and unclassified variants in seven Usher syndrome causing genes. Hum Mutat. 2008;29:E76-E87. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in RCA: 28] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 79. | Becirovic E, Ebermann I, Nagy D, Zrenner E, Seeliger MW, Bolz HJ. Usher syndrome type 1 due to missense mutations on both CDH23 alleles: investigation of mRNA splicing. Hum Mutat. 2008;29:452. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 80. | Schultz JM, Bhatti R, Madeo AC, Turriff A, Muskett JA, Zalewski CK, King KA, Ahmed ZM, Riazuddin S, Ahmad N, Hussain Z, Qasim M, Kahn SN, Meltzer MR, Liu XZ, Munisamy M, Ghosh M, Rehm HL, Tsilou ET, Griffith AJ, Zein WM, Brewer CC, Friedman TB. Allelic hierarchy of CDH23 mutations causing non-syndromic deafness DFNB12 or Usher syndrome USH1D in compound heterozygotes. J Med Genet. 2011;48:767-775. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 72] [Cited by in RCA: 78] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 81. | Ahmed ZM, Riazuddin S, Bernstein SL, Ahmed Z, Khan S, Griffith AJ, Morell RJ, Friedman TB, Wilcox ER. Mutations of the protocadherin gene PCDH15 cause Usher syndrome type 1F. Am J Hum Genet. 2001;69:25-34. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 302] [Cited by in RCA: 307] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 82. | Ahmed ZM, Goodyear R, Riazuddin S, Lagziel A, Legan PK, Behra M, Burgess SM, Lilley KS, Wilcox ER, Griffith AJ, Frolenkov GI, Belyantseva IA, Richardson GP, Friedman TB. The tip-link antigen, a protein associated with the transduction complex of sensory hair cells, is protocadherin-15. J Neurosci. 2006;26:7022-7034. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 203] [Cited by in RCA: 215] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 83. | Ahmed ZM, Riazuddin S, Aye S, Ali RA, Venselaar H, Anwar S, Belyantseva PP, Qasim M, Friedman TB. Gene structure and mutant alleles of PCDH15: nonsyndromic deafness DFNB23 and type 1 Usher syndrome. Hum Genet. 2008;124:215-223. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 59] [Cited by in RCA: 72] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 84. | Chen N, Lee H, Kim AH, Liu PK, Kang EY, Tseng YJ, Seo GH, Khang R, Liu L, Chen KJ, Wu WC, Hsiao MC, Wang NK. Case report: novel PCDH15 variant causes usher syndrome type 1F with congenital hearing loss and syndromic retinitis pigmentosa. BMC Ophthalmol. 2022;22:441. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 85. | Zheng QY, Yan D, Ouyang XM, Du LL, Yu H, Chang B, Johnson KR, Liu XZ. Digenic inheritance of deafness caused by mutations in genes encoding cadherin 23 and protocadherin 15 in mice and humans. Hum Mol Genet. 2005;14:103-111. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 88] [Cited by in RCA: 101] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 86. | Weil D, El-Amraoui A, Masmoudi S, Mustapha M, Kikkawa Y, Lainé S, Delmaghani S, Adato A, Nadifi S, Zina ZB, Hamel C, Gal A, Ayadi H, Yonekawa H, Petit C. Usher syndrome type I G (USH1G) is caused by mutations in the gene encoding SANS, a protein that associates with the USH1C protein, harmonin. Hum Mol Genet. 2003;12:463-471. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 202] [Cited by in RCA: 203] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 87. | Bashir R, Fatima A, Naz S. A frameshift mutation in SANS results in atypical Usher syndrome. Clin Genet. 2010;78:601-603. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in RCA: 22] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 88. | Kalay E, de Brouwer AP, Caylan R, Nabuurs SB, Wollnik B, Karaguzel A, Heister JG, Erdol H, Cremers FP, Cremers CW, Brunner HG, Kremer H. A novel D458V mutation in the SANS PDZ binding motif causes atypical Usher syndrome. J Mol Med (Berl). 2005;83:1025-1032. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in RCA: 30] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 89. | Yildirim A, Mozaffari-Jovin S, Wallisch AK, Schäfer J, Ludwig SEJ, Urlaub H, Lührmann R, Wolfrum U. SANS (USH1G) regulates pre-mRNA splicing by mediating the intra-nuclear transfer of tri-snRNP complexes. Nucleic Acids Res. 2021;49:5845-5866. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 90. | Zein WM, Falsini B, Tsilou ET, Turriff AE, Schultz JM, Friedman TB, Brewer CC, Zalewski CK, King KA, Muskett JA, Rehman AU, Morell RJ, Griffith AJ, Sieving PA. Cone responses in Usher syndrome types 1 and 2 by microvolt electroretinography. Invest Ophthalmol Vis Sci. 2014;56:107-114. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 91. | Pater JA, Green J, O'Rielly DD, Griffin A, Squires J, Burt T, Fernandez S, Fernandez B, Houston J, Zhou J, Roslin NM, Young TL. Novel Usher syndrome pathogenic variants identified in cases with hearing and vision loss. BMC Med Genet. 2019;20:68. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 92. | Toualbi L, Toms M, Moosajee M. USH2A-retinopathy: From genetics to therapeutics. Exp Eye Res. 2020;201:108330. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in RCA: 62] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 93. | Ahmed AN, Tahir R, Khan N, Ahmad M, Dawood M, Basit A, Yasin M, Nowshid M, Marwan M, Sultan K, Saleha S. USH2A gene variants cause Keratoconus and Usher syndrome phenotypes in Pakistani families. BMC Ophthalmol. 2021;21:191. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in RCA: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 94. | Zhu T, Chen DF, Wang L, Wu S, Wei X, Li H, Jin ZB, Sui R. USH2A variants in Chinese patients with Usher syndrome type II and non-syndromic retinitis pigmentosa. Br J Ophthalmol. 2021;105:694-703. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 95. | Yan D, Liu XZ. Genetics and pathological mechanisms of Usher syndrome. J Hum Genet. 2010;55:327-335. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 117] [Cited by in RCA: 132] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 96. | Ng TK, Tang W, Cao Y, Chen S, Zheng Y, Xiao X, Chen H. Whole exome sequencing identifies novel USH2A mutations and confirms Usher syndrome 2 diagnosis in Chinese retinitis pigmentosa patients. Sci Rep. 2019;9:5628. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 97. | Daich Varela M, Wong SW, Kiray G, Schlottmann PG, Arno G, Shams ANA, Mahroo OA, Webster AR, AlTalbishi A, Michaelides M. Detailed Clinical, Ophthalmic, and Genetic Characterization of ADGRV1-Associated Usher Syndrome. Am J Ophthalmol. 2023;256:186-195. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 2] [Reference Citation Analysis (0)] |
| 98. | Stemerdink M, García-Bohórquez B, Schellens R, Garcia-Garcia G, Van Wijk E, Millan JM. Genetics, pathogenesis and therapeutic developments for Usher syndrome type 2. Hum Genet. 2022;141:737-758. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 99. | Linnert J, Knapp B, Güler BE, Boldt K, Ueffing M, Wolfrum U. Usher syndrome proteins ADGRV1 (USH2C) and CIB2 (USH1J) interact and share a common interactome containing TRiC/CCT-BBS chaperonins. Front Cell Dev Biol. 2023;11:1199069. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 100. | Guan Y, Du HB, Yang Z, Wang YZ, Ren R, Liu WW, Zhang C, Zhang JH, An WT, Li NN, Zeng XX, Li J, Sun YX, Wang YF, Yang F, Yang J, Xiong W, Yu X, Chai RJ, Tu XM, Sun JP, Xu ZG. Deafness-Associated ADGRV1 Mutation Impairs USH2A Stability through Improper Phosphorylation of WHRN and WDSUB1 Recruitment. Adv Sci (Weinh). 2023;10:e2205993. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 101. | Audo I, Bujakowska K, Mohand-Saïd S, Tronche S, Lancelot ME, Antonio A, Germain A, Lonjou C, Carpentier W, Sahel JA, Bhattacharya S, Zeitz C. A novel DFNB31 mutation associated with Usher type 2 syndrome showing variable degrees of auditory loss in a consanguineous Portuguese family. Mol Vis. 2011;17:1598-1606. [PubMed] [Cited in This Article: ] |
| 102. | Wang L, Zou J, Shen Z, Song E, Yang J. Whirlin interacts with espin and modulates its actin-regulatory function: an insight into the mechanism of Usher syndrome type II. Hum Mol Genet. 2012;21:692-710. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in RCA: 48] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 103. | Mburu P, Mustapha M, Varela A, Weil D, El-Amraoui A, Holme RH, Rump A, Hardisty RE, Blanchard S, Coimbra RS, Perfettini I, Parkinson N, Mallon AM, Glenister P, Rogers MJ, Paige AJ, Moir L, Clay J, Rosenthal A, Liu XZ, Blanco G, Steel KP, Petit C, Brown SD. Defects in whirlin, a PDZ domain molecule involved in stereocilia elongation, cause deafness in the whirler mouse and families with DFNB31. Nat Genet. 2003;34:421-428. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 244] [Cited by in RCA: 249] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 104. | Aller E, Jaijo T, van Wijk E, Ebermann I, Kersten F, García-García G, Voesenek K, Aparisi MJ, Hoefsloot L, Cremers C, Díaz-Llopis M, Pennings R, Bolz HJ, Kremer H, Millán JM. Sequence variants of the DFNB31 gene among Usher syndrome patients of diverse origin. Mol Vis. 2010;16:495-500. [PubMed] [Cited in This Article: ] |
| 105. | Roberts L, George S, Greenberg J, Ramesar RS. A Founder Mutation in MYO7A Underlies a Significant Proportion of Usher Syndrome in Indigenous South Africans: Implications for the African Diaspora. Invest Ophthalmol Vis Sci. 2015;56:6671-6678. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in RCA: 8] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 106. | Marouf A, Johnson B, Alagramam KN. Usher syndrome IIIA: a review of the disorder and preclinical research advances in therapeutic approaches. Hum Genet. 2022;141:759-783. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 107. | Xu L, Bolch SN, Santiago CP, Dyka FM, Akil O, Lobanova ES, Wang Y, Martemyanov KA, Hauswirth WW, Smith WC, Handa JT, Blackshaw S, Ash JD, Dinculescu A. Clarin-1 expression in adult mouse and human retina highlights a role of Müller glia in Usher syndrome. J Pathol. 2020;250:195-204. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in RCA: 16] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 108. | Li T, Feng Y, Liu Y, He C, Liu J, Chen H, Deng Y, Li M, Li W, Song J, Niu Z, Sang S, Wen J, Men M, Chen X, Li J, Liu X, Ling J. A novel ABHD12 nonsense variant in Usher syndrome type 3 family with genotype-phenotype spectrum review. Gene. 2019;704:113-120. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in RCA: 12] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 109. | Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819-823. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 10336] [Cited by in RCA: 10977] [Article Influence: 914.8] [Reference Citation Analysis (0)] |
| 110. | Fuster-García C, García-García G, González-Romero E, Jaijo T, Sequedo MD, Ayuso C, Vázquez-Manrique RP, Millán JM, Aller E. USH2A Gene Editing Using the CRISPR System. Mol Ther Nucleic Acids. 2017;8:529-541. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in RCA: 45] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 111. | Overlack N, Goldmann T, Wolfrum U, Nagel-Wolfrum K. Gene repair of an Usher syndrome causing mutation by zinc-finger nuclease mediated homologous recombination. Invest Ophthalmol Vis Sci. 2012;53:4140-4146. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in RCA: 35] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 112. | Trouillet A, Dubus E, Dégardin J, Estivalet A, Ivkovic I, Godefroy D, García-Ayuso D, Simonutti M, Sahly I, Sahel JA, El-Amraoui A, Petit C, Picaud S. Cone degeneration is triggered by the absence of USH1 proteins but prevented by antioxidant treatments. Sci Rep. 2018;8:1968. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited in This Article: ] [Cited by in Crossref: 22] [Cited by in RCA: 33] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 113. | Van Diepen H, Dulla K, Chan HL, Schulkens I, Beumer W, Vorthoren L, den Besten C, Buil L, Turunen J, Miao J, Broekman S, de Vrieze E, Dona M, Albert S, Wijk EV, Adamson PS. QR-421a, an antisense oligonucleotide, for the treatment of retinitis pigmentosa due to USH2A exon 13 mutations. Investig Ophthalmol Vis Sci. 2019;60:3250. [Cited in This Article: ] |
| 114. | Fry LE, McClements ME, MacLaren RE. Analysis of Pathogenic Variants Correctable With CRISPR Base Editing Among Patients With Recessive Inherited Retinal Degeneration. JAMA Ophthalmol. 2021;139:319-328. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 115. | Landrum MJ, Lee JM, Benson M, Brown G, Chao C, Chitipiralla S, Gu B, Hart J, Hoffman D, Hoover J, Jang W, Katz K, Ovetsky M, Riley G, Sethi A, Tully R, Villamarin-Salomon R, Rubinstein W, Maglott DR. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44:D862-D868. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited in This Article: ] [Cited by in Crossref: 1642] [Cited by in RCA: 1903] [Article Influence: 190.3] [Reference Citation Analysis (0)] |
| 116. | Novartis Pharmaceuticals. Safety, Tolerability and Efficacy for CGF166 in Patients with Unilateral or Bilateral Severe-to-profound Hearing Loss-Ful. [cited 10 December 2023]. Available from: https://classic.clinicaltrials.gov/ct2/show/NCT02132130. [Cited in This Article: ] |
| 117. | Cosgrove D, Zallocchi M. Usher protein functions in hair cells and photoreceptors. Int J Biochem Cell Biol. 2014;46:80-89. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in RCA: 73] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 118. | Litovsky R. Development of the auditory system. Handb Clin Neurol. 2015;129:55-72. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 75] [Cited by in RCA: 74] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 119. | Gao X, Tao Y, Lamas V, Huang M, Yeh WH, Pan B, Hu YJ, Hu JH, Thompson DB, Shu Y, Li Y, Wang H, Yang S, Xu Q, Polley DB, Liberman MC, Kong WJ, Holt JR, Chen ZY, Liu DR. Treatment of autosomal dominant hearing loss by in vivo delivery of genome editing agents. Nature. 2018;553:217-221. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited in This Article: ] [Cited by in Crossref: 306] [Cited by in RCA: 378] [Article Influence: 47.3] [Reference Citation Analysis (0)] |
| 120. | Wang L, Kempton JB, Jiang H, Jodelka FM, Brigande AM, Dumont RA, Rigo F, Lentz JJ, Hastings ML, Brigande JV. Fetal antisense oligonucleotide therapy for congenital deafness and vestibular dysfunction. Nucleic Acids Res. 2020;48:5065-5080. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited in This Article: ] [Cited by in Crossref: 19] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 121. | Lentz JJ, Pan B, Ponnath A, Tran CM, Nist-Lund C, Galvin A, Goldberg H, Robillard KN, Jodelka FM, Farris HE, Huang J, Chen T, Zhu H, Zhou W, Rigo F, Hastings ML, Géléoc GSG. Direct Delivery of Antisense Oligonucleotides to the Middle and Inner Ear Improves Hearing and Balance in Usher Mice. Mol Ther. 2020;28:2662-2676. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in RCA: 27] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 122. | Hastings ML, Jones TA. Antisense Oligonucleotides for the Treatment of Inner Ear Dysfunction. Neurotherapeutics. 2019;16:348-359. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in RCA: 12] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 123. | Ponnath A, Depreux FF, Jodelka FM, Rigo F, Farris HE, Hastings ML, Lentz JJ. Rescue of Outer Hair Cells with Antisense Oligonucleotides in Usher Mice Is Dependent on Age of Treatment. J Assoc Res Otolaryngol. 2018;19:1-16. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in RCA: 29] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 124. | Nayerossadat N, Maedeh T, Ali PA. Viral and nonviral delivery systems for gene delivery. Adv Biomed Res. 2012;1:27. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited in This Article: ] [Cited by in Crossref: 591] [Cited by in RCA: 540] [Article Influence: 41.5] [Reference Citation Analysis (0)] |
| 125. | Ivanchenko MV, Hanlon KS, Hathaway DM, Klein AJ, Peters CW, Li Y, Tamvakologos PI, Nammour J, Maguire CA, Corey DP. AAV-S: A versatile capsid variant for transduction of mouse and primate inner ear. Mol Ther Methods Clin Dev. 2021;21:382-398. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in RCA: 37] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 126. | Allocca M, Doria M, Petrillo M, Colella P, Garcia-Hoyos M, Gibbs D, Kim SR, Maguire A, Rex TS, Di Vicino U, Cutillo L, Sparrow JR, Williams DS, Bennett J, Auricchio A. Serotype-dependent packaging of large genes in adeno-associated viral vectors results in effective gene delivery in mice. J Clin Invest. 2008;118:1955-1964. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 196] [Cited by in RCA: 221] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 127. | Rodrigues GA, Shalaev E, Karami TK, Cunningham J, Slater NKH, Rivers HM. Pharmaceutical Development of AAV-Based Gene Therapy Products for the Eye. Pharm Res. 2018;36:29. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited in This Article: ] [Cited by in Crossref: 95] [Cited by in RCA: 148] [Article Influence: 21.1] [Reference Citation Analysis (0)] |
| 128. | DiCarlo JE, Mahajan VB, Tsang SH. Gene therapy and genome surgery in the retina. J Clin Invest. 2018;128:2177-2188. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 78] [Cited by in RCA: 95] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 129. | Li T, Adamian M, Roof DJ, Berson EL, Dryja TP, Roessler BJ, Davidson BL. In vivo transfer of a reporter gene to the retina mediated by an adenoviral vector. Invest Ophthalmol Vis Sci. 1994;35:2543-2549. [PubMed] [Cited in This Article: ] |
| 130. | Hashimoto T, Gibbs D, Lillo C, Azarian SM, Legacki E, Zhang XM, Yang XJ, Williams DS. Lentiviral gene replacement therapy of retinas in a mouse model for Usher syndrome type 1B. Gene Ther. 2007;14:584-594. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 76] [Cited by in RCA: 90] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 131. | Zallocchi M, Binley K, Lad Y, Ellis S, Widdowson P, Iqball S, Scripps V, Kelleher M, Loader J, Miskin J, Peng YW, Wang WM, Cheung L, Delimont D, Mitrophanous KA, Cosgrove D. EIAV-based retinal gene therapy in the shaker1 mouse model for usher syndrome type 1B: development of UshStat. PLoS One. 2014;9:e94272. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited in This Article: ] [Cited by in Crossref: 70] [Cited by in RCA: 79] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 132. | Emptoz A, Michel V, Lelli A, Akil O, Boutet de Monvel J, Lahlou G, Meyer A, Dupont T, Nouaille S, Ey E, Franca de Barros F, Beraneck M, Dulon D, Hardelin JP, Lustig L, Avan P, Petit C, Safieddine S. Local gene therapy durably restores vestibular function in a mouse model of Usher syndrome type 1G. Proc Natl Acad Sci U S A. 2017;114:9695-9700. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 74] [Cited by in RCA: 88] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 133. | Zou J, Luo L, Shen Z, Chiodo VA, Ambati BK, Hauswirth WW, Yang J. Whirlin replacement restores the formation of the USH2 protein complex in whirlin knockout photoreceptors. Invest Ophthalmol Vis Sci. 2011;52:2343-2351. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 46] [Cited by in RCA: 52] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 134. | Hordeaux J, Wang Q, Katz N, Buza EL, Bell P, Wilson JM. The Neurotropic Properties of AAV-PHP.B Are Limited to C57BL/6J Mice. Mol Ther. 2018;26:664-668. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 212] [Cited by in RCA: 289] [Article Influence: 41.3] [Reference Citation Analysis (0)] |
| 135. | Trapani I, Colella P, Sommella A, Iodice C, Cesi G, de Simone S, Marrocco E, Rossi S, Giunti M, Palfi A, Farrar GJ, Polishchuk R, Auricchio A. Effective delivery of large genes to the retina by dual AAV vectors. EMBO Mol Med. 2014;6:194-211. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited in This Article: ] [Cited by in Crossref: 188] [Cited by in RCA: 190] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 136. | Maeda Y, Fukushima K, Nishizaki K, Smith RJ. In vitro and in vivo suppression of GJB2 expression by RNA interference. Hum Mol Genet. 2005;14:1641-1650. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 82] [Cited by in RCA: 91] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 137. | Shibata SB, Ranum PT, Moteki H, Pan B, Goodwin AT, Goodman SS, Abbas PJ, Holt JR, Smith RJH. RNA Interference Prevents Autosomal-Dominant Hearing Loss. Am J Hum Genet. 2016;98:1101-1113. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited in This Article: ] [Cited by in Crossref: 72] [Cited by in RCA: 85] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 138. | Goyal N, Narayanaswami P. Making sense of antisense oligonucleotides: A narrative review. Muscle Nerve. 2018;57:356-370. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 28] [Cited by in RCA: 30] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 139. | Keeling KM, Wang D, Conard SE, Bedwell DM. Suppression of premature termination codons as a therapeutic approach. Crit Rev Biochem Mol Biol. 2012;47:444-463. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 86] [Cited by in RCA: 80] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 140. | Selimoglu E. Aminoglycoside-induced ototoxicity. Curr Pharm Des. 2007;13:119-126. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 258] [Cited by in RCA: 270] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 141. | Mingeot-Leclercq MP, Tulkens PM. Aminoglycosides: nephrotoxicity. Antimicrob Agents Chemother. 1999;43:1003-1012. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 578] [Cited by in RCA: 568] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 142. | Nudelman I, Rebibo-Sabbah A, Shallom-Shezifi D, Hainrichson M, Stahl I, Ben-Yosef T, Baasov T. Redesign of aminoglycosides for treatment of human genetic diseases caused by premature stop mutations. Bioorg Med Chem Lett. 2006;16:6310-6315. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 58] [Cited by in RCA: 50] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 143. | Neuhaus C, Eisenberger T, Decker C, Nagl S, Blank C, Pfister M, Kennerknecht I, Müller-Hofstede C, Charbel Issa P, Heller R, Beck B, Rüther K, Mitter D, Rohrschneider K, Steinhauer U, Korbmacher HM, Huhle D, Elsayed SM, Taha HM, Baig SM, Stöhr H, Preising M, Markus S, Moeller F, Lorenz B, Nagel-Wolfrum K, Khan AO, Bolz HJ. Next-generation sequencing reveals the mutational landscape of clinically diagnosed Usher syndrome: copy number variations, phenocopies, a predominant target for translational read-through, and PEX26 mutated in Heimler syndrome. Mol Genet Genomic Med. 2017;5:531-552. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited in This Article: ] [Cited by in Crossref: 40] [Cited by in RCA: 50] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 144. | Goldmann T, Overlack N, Wolfrum U, Nagel-Wolfrum K. PTC124-mediated translational readthrough of a nonsense mutation causing Usher syndrome type 1C. Hum Gene Ther. 2011;22:537-547. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 76] [Cited by in RCA: 71] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 145. | Goldmann T, Overlack N, Möller F, Belakhov V, van Wyk M, Baasov T, Wolfrum U, Nagel-Wolfrum K. A comparative evaluation of NB30, NB54 and PTC124 in translational read-through efficacy for treatment of an USH1C nonsense mutation. EMBO Mol Med. 2012;4:1186-1199. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited in This Article: ] [Cited by in Crossref: 75] [Cited by in RCA: 81] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 146. | Namgoong JH, Bertoni C. Clinical potential of ataluren in the treatment of Duchenne muscular dystrophy. Degener Neurol Neuromuscul Dis. 2016;6:37-48. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in RCA: 13] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 147. | Lahlou G, Daoudi H, Ferrary E, Jia H, De Bergh M, Nguyen Y, Sterkers O, Mosnier I. Candidacy for Cochlear Implantation in Prelingual Profoundly Deaf Adult Patients. J Clin Med. 2022;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in RCA: 5] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 148. | Glennon E, Svirsky MA, Froemke RC. Auditory cortical plasticity in cochlear implant users. Curr Opin Neurobiol. 2020;60:108-114. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in RCA: 44] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 149. | Davies C, Bergman J, Misztal C, Ramchandran R, Mittal J, Bulut E, Shah V, Mittal R, Eshraghi AA. The Outcomes of Cochlear Implantation in Usher Syndrome: A Systematic Review. J Clin Med. 2021;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 150. | Jatana KR, Thomas D, Weber L, Mets MB, Silverman JB, Young NM. Usher syndrome: characteristics and outcomes of pediatric cochlear implant recipients. Otol Neurotol. 2013;34:484-489. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in RCA: 24] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 151. | Henricson C, Wass M, Lidestam B, Möller C, Lyxell B. Cognitive skills in children with Usher syndrome type 1 and cochlear implants. Int J Pediatr Otorhinolaryngol. 2012;76:1449-1457. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 152. | Sharma A, Dorman MF, Spahr AJ. Rapid development of cortical auditory evoked potentials after early cochlear implantation. Neuroreport. 2002;13:1365-1368. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 133] [Cited by in RCA: 151] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 153. | Chweya CM, May MM, DeJong MD, Baas BS, Lohse CM, Driscoll CLW, Carlson ML. Language and Audiological Outcomes Among Infants Implanted Before 9 and 12 Months of Age Versus Older Children: A Continuum of Benefit Associated With Cochlear Implantation at Successively Younger Ages. Otol Neurotol. 2021;42:686-693. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in RCA: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 154. | Wu SS, Sbeih F, Anne S, Cohen MS, Schwartz S, Liu YC, Appachi S. Auditory Outcomes in Children Who Undergo Cochlear Implantation Before 12 Months of Age: A Systematic Review. Otolaryngol Head Neck Surg. 2023;169:210-220. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 155. | Remjasz-Jurek A, Clarós P, Clarós-Pujol A, Pujol C, Clarós A. Outcomes of cochlear implantation in children with Usher syndrome: a long-term observation. Eur Arch Otorhinolaryngol. 2023;280:2119-2132. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 156. | Damen GW, Pennings RJ, Snik AF, Mylanus EA. Quality of life and cochlear implantation in Usher syndrome type I. Laryngoscope. 2006;116:723-728. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 34] [Cited by in RCA: 43] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 157. | Geers AE, Nicholas JG. Enduring advantages of early cochlear implantation for spoken language development. J Speech Lang Hear Res. 2013;56:643-655. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 126] [Cited by in RCA: 134] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 158. | Colletti L, Mandalà M, Zoccante L, Shannon RV, Colletti V. Infants versus older children fitted with cochlear implants: performance over 10 years. Int J Pediatr Otorhinolaryngol. 2011;75:504-509. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 87] [Cited by in RCA: 88] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 159. | Simon F, Roman S, Truy E, Barone P, Belmin J, Blanchet C, Borel S, Charpiot A, Coez A, Deguine O, Farinetti A, Godey B, Lazard D, Marx M, Mosnier I, Nguyen Y, Teissier N, Virole B, Lescanne E, Loundon N. Guidelines (short version) of the French Society of Otorhinolaryngology (SFORL) on pediatric cochlear implant indications. Eur Ann Otorhinolaryngol Head Neck Dis. 2019;136:385-391. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in RCA: 7] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 160. | Lammers MJW, Versnel H, Topsakal V, van Zanten GA, Grolman W. Predicting Performance and Non-Use in Prelingually Deaf and Late-Implanted Cochlear Implant Users. Otol Neurotol. 2018;39:e436-e442. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 8] [Cited by in RCA: 15] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 161. | Debruyne J, Janssen M, Brokx J. Late Cochlear Implantation in Early-Deafened Adults: A Detailed Analysis of Auditory and Self-Perceived Benefits. Audiol Neurootol. 2017;22:364-376. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in RCA: 13] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 162. | Bosco E, Nicastri M, Ballantyne D, Viccaro M, Ruoppolo G, Ionescu Maddalena A, Mancini P. Long term results in late implanted adolescent and adult CI recipients. Eur Arch Otorhinolaryngol. 2013;270:2611-2620. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in RCA: 18] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 163. | Hoshino AC, Echegoyen A, Goffi-Gomez MV, Tsuji RK, Bento RF. Outcomes of Late Implantation in Usher Syndrome Patients. Int Arch Otorhinolaryngol. 2017;21:140-143. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in RCA: 11] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 164. | Pietola L, Aarnisalo AA, Abdel-Rahman A, Västinsalo H, Isosomppi J, Löppönen H, Kentala E, Johansson R, Valtonen H, Vasama JP, Sankila EM, Jero J. Speech recognition and communication outcomes with cochlear implantation in Usher syndrome type 3. Otol Neurotol. 2012;33:38-41. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 12] [Cited by in RCA: 14] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 165. | Koffler T, Ushakov K, Avraham KB. Genetics of Hearing Loss: Syndromic. Otolaryngol Clin North Am. 2015;48:1041-1061. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 87] [Cited by in RCA: 101] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 166. | Liu XZ, Angeli SI, Rajput K, Yan D, Hodges AV, Eshraghi A, Telischi FF, Balkany TJ. Cochlear implantation in individuals with Usher type 1 syndrome. Int J Pediatr Otorhinolaryngol. 2008;72:841-847. [RCA] [PubMed] [DOI] [Full Text] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in RCA: 43] [Article Influence: 2.5] [Reference Citation Analysis (0)] |