Copyright
©The Author(s) 2016.
World J Clin Oncol. Apr 10, 2016; 7(2): 135-148
Published online Apr 10, 2016. doi: 10.5306/wjco.v7.i2.135
Published online Apr 10, 2016. doi: 10.5306/wjco.v7.i2.135
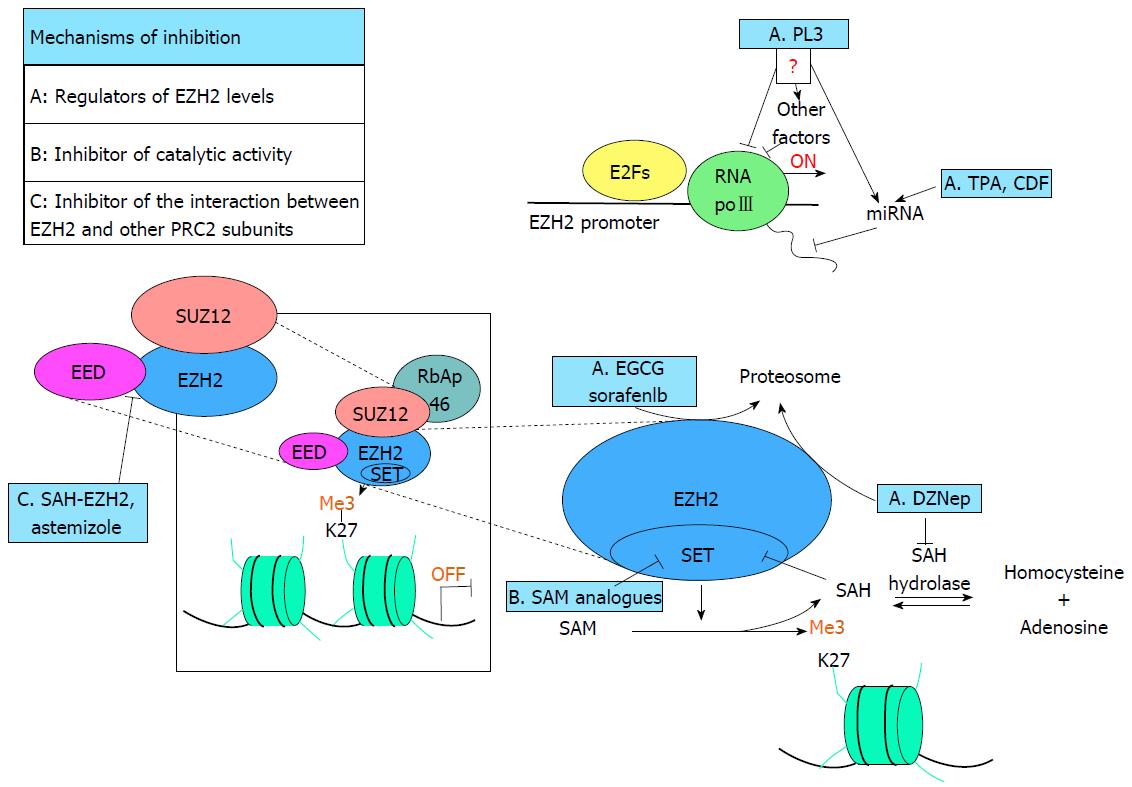
Figure 1 Schematic representation of polycomb-repressive complex 2 inhibition mechanisms.
The image enclosed in the square represent the PRC2 active complex, able to methylate the lysine 27 of the histone H3 (H3K27me3), suppressing the transcription of target genes. Dashed lines pointed a part of the complex or the methylation pathway. The main PRC2 inhibitors (blue square) are included in the figure and labeled with the letters A, B or C depending on the mechanism of action (drugs marked with A are responsible for EZH2 depletion, with B are inhibitors of the catalytic activity of EZH2 and with C are able to destroy PRC2 complex. In group A PL3 is able to repress EZH2 expression but is not clear if the mechanism is direct or involves other factors or miRNAs. TPA and CDF reduce levels of EZH2 inducing the expression of some miRNA. EGCG, Sorafenib and DZNep promote proteasome-dependent EZH2 degradation. On the other hand DZNep, inhibiting SAH-Hydrolase, leads to an accumulation of SAH that, in turn, hampers try-methylation of H3K27me3 inhibiting EZH2 SET domain. Group B, SAM analogues compete with SAM for the binding with the substrate pocket blocking SET domain activity. Finally in the group C both the peptide SAH-EZH2 and Astemizole destroy the binding between EZH2 and EED event required for the activity of PRC2 complex. SAM: S-adenosylmethionine; SAH: S-adenosylhomocysteine; PRC2: Polycomb-repressive complex 2; EZH2: Enhancer of Zeste Homolog 2; EGCG: (-)-epigallocatechin-3-gallate; DZNep: 3-Deazaneplanocin A; SAH-EZH2: Stabilized alpha-helix of EZH2.
- Citation: Marchesi I, Bagella L. Targeting Enhancer of Zeste Homolog 2 as a promising strategy for cancer treatment. World J Clin Oncol 2016; 7(2): 135-148
- URL: https://www.wjgnet.com/2218-4333/full/v7/i2/135.htm
- DOI: https://dx.doi.org/10.5306/wjco.v7.i2.135