Copyright
©The Author(s) 2024.
World J Clin Oncol. Aug 24, 2024; 15(8): 1061-1077
Published online Aug 24, 2024. doi: 10.5306/wjco.v15.i8.1061
Published online Aug 24, 2024. doi: 10.5306/wjco.v15.i8.1061
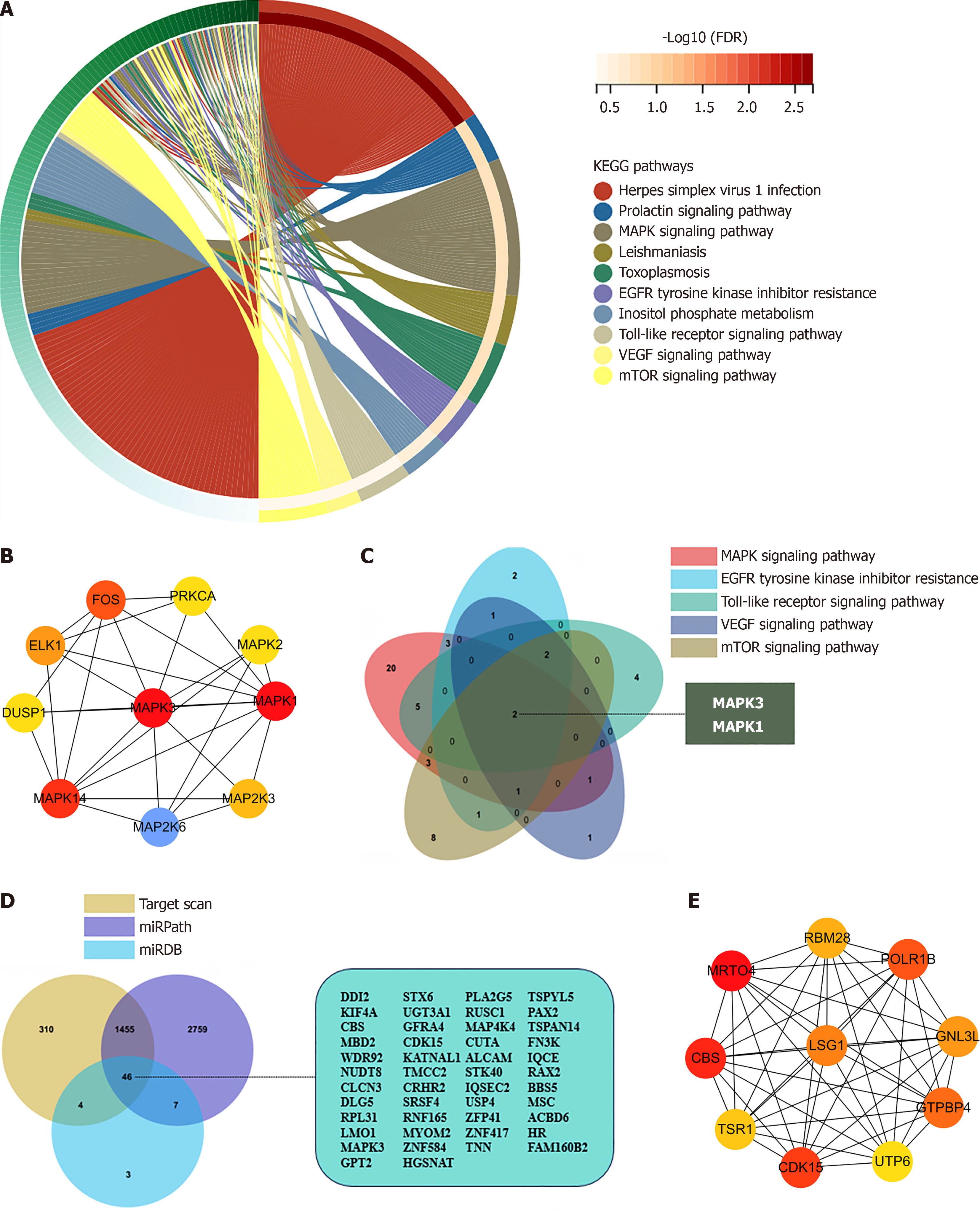
Figure 3 The bioinformatics analysis of the target genes of exosome hsa-miR-483-5p.
A: Represents the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways enrichment results of target genes; B: The protein-protein interaction (PPI) network and the hub genes selected in the enriched five KEGG pathways; C: The selected common target genes of hsa-miR-483-5p enriched in the five KEGG pathways, with two identified hub genes-MAPK1 and MAPK3; D: The PPI network and the hub genes selected from the target genes of exosome hsa-miR-483-5p; E: The selection of target genes of hsa-miR-483-5p from the three miRNA-associated databases, with chosen 46 common genes. KEGG: Kyoto Encyclopedia of Genes and Genomes; PPI: Protein-protein interaction.
- Citation: Li GB, Shi WK, Zhang X, Qiu XY, Lin GL. Hsa-miR-483-5p/mRNA network that regulates chemotherapy resistance in locally advanced rectal cancer identified through plasma exosome transcriptomics. World J Clin Oncol 2024; 15(8): 1061-1077
- URL: https://www.wjgnet.com/2218-4333/full/v15/i8/1061.htm
- DOI: https://dx.doi.org/10.5306/wjco.v15.i8.1061