Copyright
©The Author(s) 2017.
World J Biol Chem. Feb 26, 2017; 8(1): 32-44
Published online Feb 26, 2017. doi: 10.4331/wjbc.v8.i1.32
Published online Feb 26, 2017. doi: 10.4331/wjbc.v8.i1.32
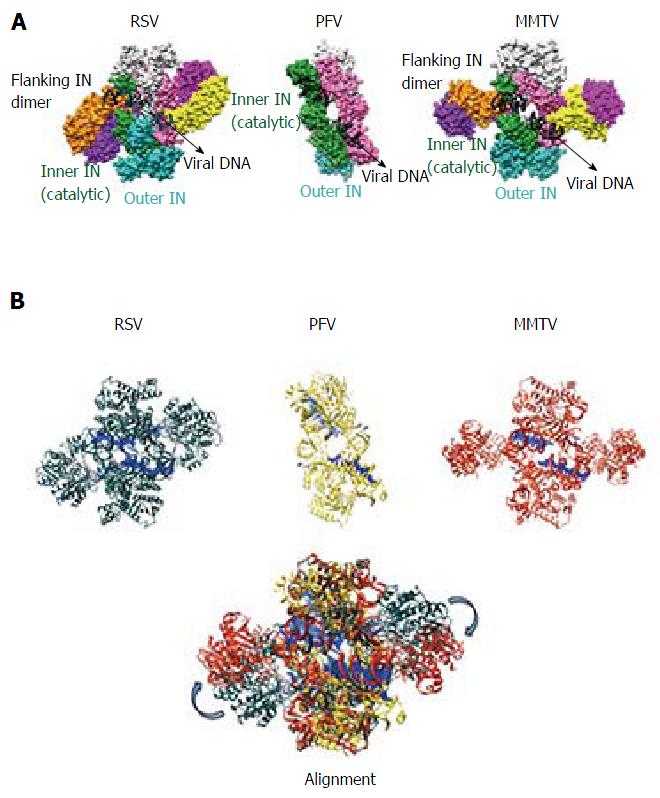
Figure 3 Comparison of prototype foamy virus, Rous sarcoma virus and mouse mammary tumor virus intasomes.
A: The PFV intasome comprises two catalytic inner subunits (green and pink) and two outer supportive INs (cyan and light grey). Only the CCDs of the outer subunits are discernable in crystallographic electron density maps. RSV and MMTV share the PFV intasome core architecture and employ two additional flanking IN dimers (orange-purple and yellow-dark pink) to complete the intasome structures; B: Three-dimensional alignment of RSV (grey, PDB accession code 5EJK), PFV (yellow, PDB code 3L2Q), and MMTV (red, PDB code 3JCA) intasome structures was performed using Chimera. For the MMTV intasome, flanking dimers were unambiguously positioned into the intasome core of the cryo-EM map via rigid-body docking. The alignment reveals a high degree of flexibility (approximately 30-40 Å) for the flanking RSV and MMTV dimers relative to the common intasome core structures (arrows). PFV: Prototype foamy virus; RSV: Rous sarcoma virus; MMTV: Mouse mammary tumor virus.
- Citation: Grawenhoff J, Engelman AN. Retroviral integrase protein and intasome nucleoprotein complex structures. World J Biol Chem 2017; 8(1): 32-44
- URL: https://www.wjgnet.com/1949-8454/full/v8/i1/32.htm
- DOI: https://dx.doi.org/10.4331/wjbc.v8.i1.32