Copyright
©The Author(s) 2017.
World J Biol Chem. Feb 26, 2017; 8(1): 32-44
Published online Feb 26, 2017. doi: 10.4331/wjbc.v8.i1.32
Published online Feb 26, 2017. doi: 10.4331/wjbc.v8.i1.32
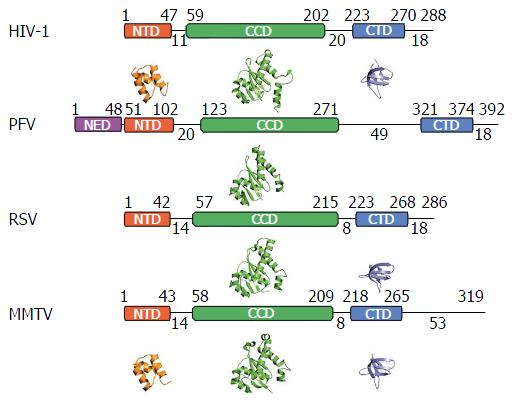
Figure 2 Integrase domain organization and representative secondary structures.
Starting and ending residues for integrase (IN) domains are indicated above the boxes, and interdomain linker lengths as well as C-terminal tail lengths are indicated below the lines. Crystal structures of N-terminal domains (NTDs), catalytic core domains (CCDs), and C-terminal domains (CTDs) are provided underneath the corresponding schematic IN representation. Crystal structures in the absence of DNA are not available for the PFV NTD extension domain (NED), NTD, or CTD, as well as for the RSV NTD. PDB accession codes: HIV-1 (NTD, 1K6Y; CCD, 1BIU; CTD, 1EX4), PFV (CCD, 3DLR), RSV (CCD, 1C0M; CTD, 1C0M) and MMTV (NTD, 5CZ2; CCD, 5CZ1; CTD, 5D7U). HIV: Human immunodeficiency virus; PFV: Prototype foamy virus; RSV: Rous sarcoma virus; MMTV: Mouse mammary tumor virus.
- Citation: Grawenhoff J, Engelman AN. Retroviral integrase protein and intasome nucleoprotein complex structures. World J Biol Chem 2017; 8(1): 32-44
- URL: https://www.wjgnet.com/1949-8454/full/v8/i1/32.htm
- DOI: https://dx.doi.org/10.4331/wjbc.v8.i1.32