Copyright
©The Author(s) 2016.
World J Biol Chem. Feb 26, 2016; 7(1): 110-127
Published online Feb 26, 2016. doi: 10.4331/wjbc.v7.i1.110
Published online Feb 26, 2016. doi: 10.4331/wjbc.v7.i1.110
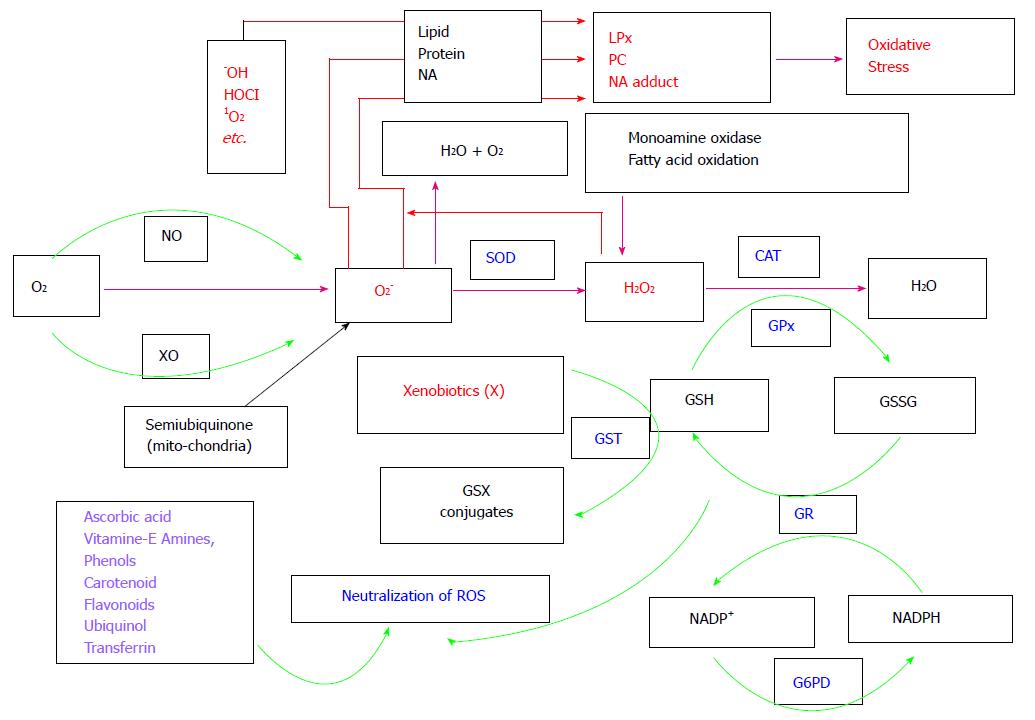
Figure 1 Pathways of active oxygen species metabolism.
OS physiology starts with O2 consumption in animals. O2 is incompletely reduced to super oxide radicals (O2-) via NO or XO or in mitochondria due incomplete reduction. O2- is dismutated to H2O2 by enzyme SOD. H2O2 is then scavenged by either CAT, or glutathione peroxidase with the help of the reduced glutathione (GSH, which form oxidised glutathione, i.e., GSSG). GSSG is recycled to GSH by the enzyme GR with the help of NADPH (which is converted into NADP+). NADP+ is reduced back to NADPH by the enzyme G6PD. GST is also responsible to remove xenobiotics (which are responsible to produce ROS) from cells with the help of GSH. Peroxiredoxins and thioredoxins system and glutaredoxins are responsible for scavenging H2O2 and reduction of other proteins (not shown in Figure). Redox regulatory non-enzymatic molecules such as ascorbic acid, flavonoids and phenols can also remove ROS such as OH, HOCL, O2, O2- and H2O2. With insufficient antioxidant defence, more ROS accumulation in cells occurs and it leads to oxidation of biomolecules such as proteins, lipids and nucleic acids to form PC, LPx and NA adducts, respectively. Formation of LPx, PC and NA leads to a disorder condition called as oxidative stress. OS: Oxidative stress; NO: Nitric oxidase; XO: Xanthine oxidase; SOD: Superoxide dismutase; CAT: Catalase; GR: Glutathione reductase; G6PD: Glucose-6-phosphate dehydrogenase; GST: Glutathione-S-transferase; PC: Protein carbonyls; LPx: Lipid peroxides; NA: Nucleic acid.
- Citation: Paital B, Panda SK, Hati AK, Mohanty B, Mohapatra MK, Kanungo S, Chainy GBN. Longevity of animals under reactive oxygen species stress and disease susceptibility due to global warming. World J Biol Chem 2016; 7(1): 110-127
- URL: https://www.wjgnet.com/1949-8454/full/v7/i1/110.htm
- DOI: https://dx.doi.org/10.4331/wjbc.v7.i1.110