Copyright
©The Author(s) 2015.
World J Biol Chem. Nov 26, 2015; 6(4): 366-378
Published online Nov 26, 2015. doi: 10.4331/wjbc.v6.i4.366
Published online Nov 26, 2015. doi: 10.4331/wjbc.v6.i4.366
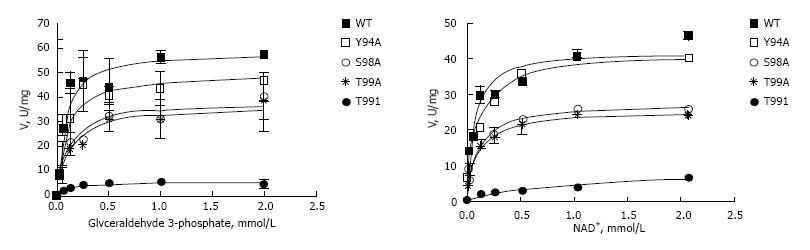
Figure 4 Kinetic analysis of wild type and mutated forms of Hisx6-tagged glyceraldehyde 3-phosphate dehydrogenase prepared in BL21 (DE3) Escherichia coli expression system.
Conditions for the glycolytic assay are indicated in “Materials and Methods”. A: Reaction was performed with 0.26 mmol/L NAD+ and varying concentrations of D-glyceraldehyde-3-phosphate (0-2 mmol/L); B: Reaction was performed with 0.51 mmol/L D-glyceraldehyde-3-phosphate and varying concentrations of NAD+ (0-2 mmol/L). Vmax and Km were calculated by non-linear regression analysis with GraphPad Prizm 4.0 as described in Materials and Methods. GAPDH: Glyceraldehyde 3-phosphate dehydrogenase.
- Citation: Phadke M, Krynetskaia N, Mishra A, Barrero C, Merali S, Gothe SA, Krynetskiy E. Disruption of NAD+ binding site in glyceraldehyde 3-phosphate dehydrogenase affects its intranuclear interactions. World J Biol Chem 2015; 6(4): 366-378
- URL: https://www.wjgnet.com/1949-8454/full/v6/i4/366.htm
- DOI: https://dx.doi.org/10.4331/wjbc.v6.i4.366