Copyright
©The Author(s) 2015.
World J Biol Chem. Aug 26, 2015; 6(3): 249-264
Published online Aug 26, 2015. doi: 10.4331/wjbc.v6.i3.249
Published online Aug 26, 2015. doi: 10.4331/wjbc.v6.i3.249
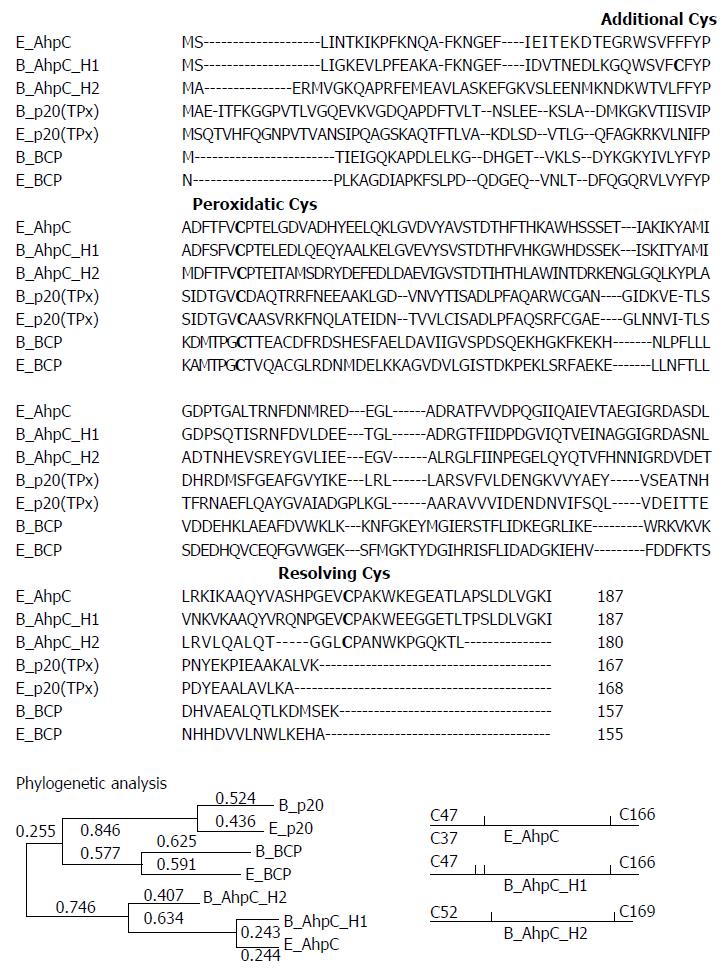
Figure 1 Amino acid sequence alignment of Bacillus subtilis and Escherichia coli peroxiredoxins.
B. subtilis database was searched using amino acid sequences around conserved N-terminal Cys residues as a query. In addition to reported bacterial genes encoding BCP and TPx enzymes, two putative AhpC-encoding genes were identified, and four B. subtilis and three E. coli Prx sequences were aligned. The two conserved Cys residues, N-terminal peroxidatic and C-terminal resolving Cys, among the AhpC proteins are shown as shaded bold letters. Proteins homologous to E. coli AhpC were identified using the ClustalW 2.1 multiple sequence alignment. The phylogenetic tree was generated by using MEGA version 6 The tree is drawn to scale, with branch lengths measured in the number of substitutions per site (next to the branches). The analysis involved 7 amino acid sequences. All positions containing gaps and missing data were eliminated. There were a total of 143 positions in the final dataset. Position of Cys residues in AhpC proteins is shown at the bottom right panel. E_BCP, E. coli BCP (accession number AAC75533); E_p20 (TPx), E. coli TPx (accession number EDV68400); E_AhpC, E. coli AhpC (accession NP_415138); B_ahpC_H1, B. subtilis AhpC homolog 1 (accession number BAA11268); B_AhpC_H2, B. subtilis AhpC homolog 2 (accession number WP_019258276); B_BCP, B. subtilis BCP (accession number AAC75533); and B_p20 (TPx), B. subtilis TPx (accession NP_390827).
-
Citation: Cha MK, Bae YJ, Kim KJ, Park BJ, Kim IH. Characterization of two alkyl hydroperoxide reductase C homologs alkyl hydroperoxide reductase C_H1 and alkyl hydroperoxide reductase C_H2 in
Bacillus subtilis . World J Biol Chem 2015; 6(3): 249-264 - URL: https://www.wjgnet.com/1949-8454/full/v6/i3/249.htm
- DOI: https://dx.doi.org/10.4331/wjbc.v6.i3.249